Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNA-182 Promotes Cell Migration by Targeting Programmed Cell Death 4 in Hepatocellular Carcinoma Cells
Authors Hu J, Wang Z, Wang J, Jian Y, Dai J, Wang X, Xiong W
Received 15 April 2020
Accepted for publication 17 August 2020
Published 16 September 2020 Volume 2020:13 Pages 9159—9167
DOI https://doi.org/10.2147/OTT.S258251
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Junwei Hu,1,2,* Zeyu Wang,1,* Jinjun Wang,3 Yicheng Jian,1,2 Jiarun Dai,1,2 Xiaoping Wang,1,2 Wujun Xiong4
1Department of Gastroenterology, Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital, Shanghai 201318, People’s Republic of China; 2Department of Digestive Endoscopy, Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital, Shanghai 201318, People’s Republic of China; 3Department of Gerontology, Shanghai Putuo Central Hospital Affiliated to Shanghai Traditional Chinese Medicine University, Shanghai 200062, People’s Republic of China; 4Department of Hepatology, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai 200120, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaoping Wang
Department of Gastroenterology, Shanghai University of Medicine & Health Sciences Affiliated Zhoupu Hospital, Shanghai 201318, People’s Republic of China
Tel +86 2168135590
Email [email protected]
Wujun Xiong
Department of Hepatology, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai 200120, People’s Republic of China
Email [email protected]
Purpose: Hepatocellular carcinoma (HCC) is the most common primary liver tumor and the third greatest cause of cancer-related death worldwide. Programmed cell death 4 (PDCD4) was reported as a potential tumor-suppressor in hepatocarcinogenesis. However, relatively little is known about mechanisms that regulate PDCD4 expression in HCC. The aim of the present study is to investigate the expression of PDCD4 and miR-182 in human HCC cell lines and clinical HCC specimens and determine whether PDCD4 is a direct target of miR-182 in HCC cell lines.
Materials: The expression of miR-182 and PDCD4 in human HCC cell lines and HCC tissues were examined using qRT-PCR and Western blot method. Transwell and wound healing assays were carried out to explore the influence of miR-182 on hepatoma cells migration. A luciferase reporter assay was conducted to confirm target association.
Results: In our research, we found that PDCD4 was downregulated, whereas miR-182 was upregulated in liver cancer cell lines and HCC tissues. Transwell and wound healing assays illustrated that miR-182 contributed to migration activities of liver cancer cell lines. Loss or increase of miR-182 can lead to a negative expression of PDCD4 protein level. The luciferase reporter assay showed that PDCD4 is a direct target of miR-182.
Conclusion: All these findings suggest that miR-182 may act as an oncogenic role in liver cancer cells by directly and negatively regulating expression of PDCD4.
Keywords: miR-182, migration, PDCD4, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC), the major form of the primary liver cancer, is one of the most common health problems worldwide, accounting for approximately 600 thousand deaths every year.1 To a large extend, HCC is associated with chronic viral infections of hepatitis type B or C and liver cirrhosis in China.2,3 It is the second leading cause of cancer-related deaths.4 So far, surgery and liver transplantation remain the best treatments for HCC patients; however, the overall 5-year survival rate is less than 15% for advanced HCC is dismal.5 Despite the increasing wealth of knowledge on the biology of HCC, the possible mechanism of hepatocarcinogenesis, the regulation of candidate oncogenes or antioncogenes, identification of novel diagnostic biomarkers and progression of effective therapies for HCC are still pressingly needed.
Programmed cell death 4 (PDCD4), a 64-kDa protein, was found to be localized on chromosome 10q24.6 It was originally identified as the neoplasmic transformation inhibitor in the JB6 mouse epidermal cell line model.7 Studies showed that PDCD4 interacts with eIFs, eIF4A, and eIF4G8,9 and thus subsequently inhibiting helicase activity of eIF4A and hence inhibiting translation. PDCD4 was assumed and then confirmed to act as a functional gene, which was suggested to inhibit proliferation10,11 and induces apoptosis.12 The loss or reduction of PDCD4 expression has been discovered in a variety of human cancer cells, such as primary pancreatic cancer,13 colon carcinoma14 and pregnancy-associated breast cancer.15 Furthermore, PDCD4 was reported as a potential tumor-suppressor in hepatocarcinogenesis.16,17
MicroRNAs (miRNAs) are a class of small non-coding regulatory RNAs with approximately 18–25 nucleotides in length.18 miRNAs negatively regulate gene expression and can trigger cleavage of target mRNAs or inhibit protein translation.19 Recent studies indicated that miRNAs can function as tumor suppressors by targeting endogenous oncogenes20,21 or act as tumor promoters by inhibiting cellular antioncogenes.22 What’s more, miRNAs have been linked to the development and progression of cancers.23 Several studies have shown that specific miRNAs are aberrantly expressed in malignant HCC cells or tissues compared to non-malignant hepatocytes or tissue.24,25
MiR-21 has been found to negatively regulate PDCD4 expression in breast cancer,26 colorectal cancer,27 malignant glioblastoma tumors.28 HBx repressed PDCD4 expression via the induction of miR-21 in HCC.29 Also, it was reported that miR-182 is a negative regulator of PDCD4 in lung adenocarcinoma cells and ovarian carcinomas.30,31 However, relatively little is known about mechanisms that regulate PDCD4 expression in hepatocellular carcinoma. The interaction of miR-182 and PTCD4 in HCC was never reported. Thus, the aim of this study is to explore the expression of PDCD4 and miR-182 in human HCC cell lines and clinical HCC specimens and determine whether PDCD4 is a direct target of miR-182 in HCC cell lines.
Materials and Methods
Cell Lines and Cell Culture
The three liver cancer cell lines HepG2, LM3, MHCC97-H, and the normal hepatic cell line LO2 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen Grand Island, NY, USA) containing 10% fetal bovine serum (FBS) (PAA, Australia) with 100 μg/mL penicillin/streptomycin and were incubated at 37°C in a 5%CO2 humidified incubator. 293T cells were cultured in DMEM supplemented with 10% FBS.
Clinical HCC Specimens
Three HCC tissue specimens were obtained from HCC patients who received surgery resection in Shanghai East Hospital, School of Medicine, Tongji University, Shanghai, China. None of the patient received any preoperative anticancer treatment. We also collected one normal liver tissue from liver traumatic rupture patient. All the tissues were immediately stored at −80°C until protein extraction. The study was approved by the Shanghai East Hospital Research Ethics Committee. All patients signed informed consent according to the committee’s regulation before surgery resection. The basic characteristics of patients were shown in supplemental Table S1.
RNA Extraction and qRT-PCR for miRNAs
Total RNA was extracted from the cultured cells using the TRIzol reagent (Invitrogen, Grand Island, NY, USA). The reverse transcription for miR-182 was performed using the TaqMan® MicroRNA RT Kit (Invitrogen, CA, USA). The primer for its cDNA synthesis was GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTGTG. To quantify the levels of miR-182, the forward primer 5ʹ-GGCGGTTTTGGCAATGGTAG-3ʹ and the reverse primer 5ʹ-GTGCAGGGTCCGAGGT-3ʹ were used. The PCR was performed on the real-time PCR system (ABI 7500 Fast, CA, USA) according to the manufacture’s protocol. RNU48 miRNA was chosen as the internal control. All reactions were performed in triplicate.
Western Blot Assay
Total protein was extracted from cultured cells or clinical tissues using RIPA buffer containing PMSF (1:1000). Protein concentration was detected by using a BCA protein assay kit (Ding Guo Biotechnology, Shanghai, China). Proteins were separated by SDS-PAGE and electroblotted to nitrocellulose membranes. After blocking at room temperature for one hour, the membrane was separately incubated at 4°C overnight with diluted primary antibody against PDCD4 (Epitomics, CA, USA). An antibody against β-actin (Santa Cruz, Biotechnology, CA, USA) was served as an endogenous reference.
Luciferase Reporter Assay
The full length PDCD4 3ʹ UTR cDNA fragments containing the putative miR-182 binding site was amplified and subcloned into pmir-GLO luciferase reporter vector (Applied Biosystems) Site-directed mutagenesis was performed to generate a mutant PDCD4 3ʹ UTR that contained mutations in the conserved miR-182 binding site. In the mutant 3ʹ UTR of the PDCD4, the nucleotide sequence complementary to nt 2–5 of the miR-182 binding site (TGCCAAA) was mutated to the sequence found in miR-182 (TCCGATA). For the luciferase reporter assay, cultured cells were seeded in 12-well plates at a density of 2×105 cells/well and were co-transfected with 0.2µg of pmirGLO-PDCD4 (wide type or mutant) and 3ug of the miR-182 mimics using the Lipofectamine 2000 reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s protocol. After 48 hours of incubation, the relative luciferase activity was measured using the Dual-luciferase reporter assay system (Promega, Beijing, China). Experiments were repeated in triplicate.
Wound Healing Assay
The groups of LM3 and MHCC97-H were transfected with inhibitors or mimics and cultured to confluence or near (>90%) in 6-well plates. A sterile 200ul pipette tip was used to scratch a straight line through the cell layer. Then the medium was removed and replaced with fresh medium. The cells were cultured and the images were taken 24h after scratching. Assays were repeated in triplicate.
Transwell Assay
Migration of cells through 8-μM pores was assessed using the Transwell cell culture chamber (6.5-mm diameter; Corning Costar, Tokyo, Japan). The transfected cells were then trypsinized, washed once using serum-free medium and resuspended. An amount of 1×104 cells within 100ul suspensions was added to the upper chamber of transwell. The lower chamber of the Transwell was filled with 600µl medium containing 10%FBS. After seeding, the cells were allowed to migrate at 37°C, 5% CO2 for 24h. Then, the cells on the upper surface were removed using a cotton bud. The remaining invaded cells were fixed with 95% ethanol and stained with 0.1% crystal violet for 30min at room temperature. The cell that had migrated to the lower surface of the filter membrane was counted in five random fields of under view at 100magnificationa light microscope. Experiments were repeated in triplicate.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0. Differences between results obtained under different experimental conditions were determined by Student’s t-test. All data are given as mean ± standard deviation (SD). All experiments were repeated in triplicate. P < 0.05 was considered statistically significant.
Results
PDCD4 Protein is Down-Regulated, Whereas miR-182 is Up-Regulated in Liver Cancer
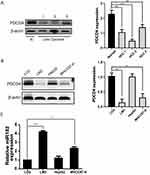
To explore whether PDCD4 is aberrantly expressed in liver cancers, we first detected the expression level of PDCD4 in 4 tissue samples, including 1 normal liver tissue and 3 HCC tissues. Western blot analysis showed that PDCD4 level was significantly lower in liver cancer samples compared to normal liver tissue (Figure 1A) (*P<0.05). Then we detected PDCD4 level in three liver cancer cell lines (HepG2, LM3, MHCC97-H) and one normal hepatic cell line LO2. According to the Western blot analysis, PDCD4 showed notably lower expression in liver cancer cell lines than in normal cells, especially in LM3 and MHCC97-H cell lines (Figure 1B) (*P<0.05, **P<0.01), and this is absolutely consistent with above result. We then analyzed miR-182 level in these four cell lines by real-time PCR, results demonstrated that miR-182 level was higher in LM3 and MHCC97-H cell lines than in normal cells (Figure 1C) (*P<0.05, **P<0.01). But the difference between normal hepatic cell line and HepG2 was insignificant. We chose LM3 and MHCC97-H cell lines for further analysis. Thus, above data indicated that PDCD4 expression is downregulated, whereas miR-182 is upregulated in human HCC cell lines and clinical HCC specimens.
miR-182 Negatively Regulated PDCD4 Expression by Targeting the PDCD4 3ʹUTR
By using the TargetScan (http://www.targetscan.org) and miRnada bioinformatics analysis, it indicated that PDCD4 3ʹUTR contains a predicted binding site for miR-182 (Figure 2A). To determine whether PDCD4 is regulated by miR-182, we conducted the luciferase reporter assays. A dual-luciferase reporter system with luciferase reporter vectors containing either the PDCD4 wide-type 3ʹUTR or mutant-type 3ʹUTR. Vectors confection with miR182 mimics significantly decreased the relative luciferase activity of the reporter containing wide-type 3ʹUTR, whereas it did not affect that of the mutant-type 3ʹUTR (Figure 2B) (*P<0.05).
Then, Western blot was applied to further validate the negative correlation between miR-182 and PDCD4. The results showed that PDCD4 expression decreased after cells transfected with miR-182 mimics in LM3 and MHCC97-H cell lines. Whereas PDCD4 expression upregulated after transfection with miR-182 inhibitor in these two cell lines (Figure 2C and D) (*P<0.05). Taken together, these data strongly demonstrated that miR-182 negatively regulates PDCD4 expression level by directly binding to its putative binding site in the 3ʹUTR.
Down-Regulation of miR-182 Restricts Cell Migration Abilities of Liver Cancer Cells
A wound healing assay was conducted to explore the role of miR-182 in regulating the migration activity of liver cancer cells. As shown in Figure 3A, inhibitor-NC group had reached a higher cell density at 24h postwounding compared to the miR-182 inhibitor group of LM3 cells. What’ more, cells transfected with mimic-NC showed a lower cell density than cultured cells transfected with mimic-miR-182 in MHCC-97H cell lines (Figure 3B).
Then, we also performed a transwell assay to further confirm the importance of miR-182 in enhancing migration activity. Panels C and D of Figure 3 showed the average numbers of cells penetrating the Transwell membrane in the three groups of LM3 and MHCC-97H cells, respectively. The average number of cells penetrating the Transwell membrane was lower in the inhibitor miR-182 group when compared with inhibitor-NC groups in LM3 cell lines (**P<0.01); additionally, the number of cells was notably higher in cells transfected with mimic-miR182 than mimic-NC of MHCC-97H cell. All these data demonstrated that inhibiting miR-182 restricts the migration capacity of liver cancer cells (*P<0.05).
Discussion
Despite the fact that great efforts have been made to overcome the difficulty in curing liver cancers, the death rate of this disease is still high.1 Thus, newly findings about molecular biology of liver cancer are urgently needed to be discovered.
PDCD4 was originally isolated from a human glioma cell. Emerging studies from various kind of human tumor confirmed that PDCD4 is a novel tumor suppressor gene and play a key role in antitumor therapies.32 Reduction of PDCD4 expression has been obtained in many cancers, such as gliomas, human salivary adenoid cystic carcinoma.33,34 Over-expression of PDCD4 suppresses tumor invasion and intravasation in many types of cancer cells such as breast, colon, liver, gastric, and ovarian cancer.35 Our research proved that PDCD4 was downregulated expression in liver cancer cell lines and liver cancer tissues, which was consistent with past researches.36 In the HepG2 cell line, the downregulation of PDCD4 was not obvious. Considering the researches reporting expression of PDCD4 relation with tumor metastasis,37 the reason account for insignificant downregulation of PDCD4 may lies in low metastatic ability of HepG2, compared with high metastatic cell lines (LM3 and MHCC97-H).38 But the detailed mechanism need to be further explored.
MiRNAs were reported to regulate expression of many human genes and to play crucial roles in a variety of processes.39–41 In the past several decades, the importance of miRNAs in liver cancer has also been gradually recognized.42,43 What’s more, increasing evidences indicate the key role of miR-182 in cancers. Previous research demonstrated that miR-182 is frequently over-expressed in numerous tumors.44,45 Increase of miR-182 was also suggested to be observed in liver cancer tissues.46,47 In our research, results also showed that the upregulation of miR-182 is a frequent event in human HCC tissues and cancer cell lines.
Above results indicated that there is a negative relation between PDCD4 and miR-182. The interaction may exist in PDCD4 and miR-182 in HCC. The luciferase reporter assay data supported our hypothesis that PDCD4 is a direct target of miR-182. And Western blot also indicated that loss of miR-182 positively upregulate the expression of PDCD4, on the contrary, overexpression of miR-182 downregulate the PDCD4 expression. Transwell and wound healing assays results illustrated that miR-182 contributed to migration activities of LM3 and MHCC-97H liver cancer cells. Past reports have showed that miR-182 is a negative regulator of PDCD4 in lung adenocarcinoma cells and ovarian carcinomas.30,31 Other researches indicated that miR-182 contributes to cell adhesion-mediated drug resistance via targeting PDCD4 in multiple myeloma.48 MicroRNA-182 modulates chemosensitivity by targeting PDCD4 in non-small cell lung cancer.49 Our research has proved that miR-182 may act as an oncogenic role in liver cancer cells by directly and negatively regulating expression of PDCD4. However, the deeper mechanism of miR-182 regulating the expression of PDCD4 needs further research.
In conclusion, our study found that miR-182 directly and negatively regulates PDCD4 and downregulation of miR-182 inhibits migration in HCC. Our findings offer a new regulatory mechanism of PDCD4 expression in HCC. This knowledge may give a new sight on the mechanism of miRNA in regulating liver cancer cells and be meaningful for the expanding of new strategies for HCC treatment.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available upon reasonable request from the corresponding author WJX.
Ethics Approval and Consent
Not applicable.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma–epidemiological trends and risk factors. Dig Dis. 2009;27(2):80–92. doi:10.1159/000218339
2. Raza SA, Clifford GM, Franceschi S. Worldwide variation in the relative importance of hepatitis B and hepatitis C viruses in hepatocellular carcinoma: a systematic review. Br J Cancer. 2007;96(7):1127–1134. doi:10.1038/sj.bjc.6603649
3. Yan SY, Fan JG, Qio L. Hepatitis B virus (HBV) infection and hepatocellular carcinoma- new insights for an old topic. Curr Cancer Drug Targets. 2017;17(6):505–511. doi:10.2174/1568009616666160926124530
4. Han Q, Zhao H, Jiang Y, Yin C, Zhang J. HCC-derived exosomes: critical player and target for cancer immune escape. Cells-Basel. 2019;8(6):558.
5. DiStefano JK, Davis B. Diagnostic and Prognostic Potential of AKR1B10 in Human Hepatocellular Carcinoma. Cancers (Basel). 2019;11. doi:10.3390/cancers11040486
6. Soejima H, Miyoshi O, Yoshinaga H, et al. Assignment of the programmed cell death 4 gene (PDCD4) to human chromosome band 10q24 by in situ hybridization. Cytogenet Cell Genet. 1999;87(1–2):113–114. doi:10.1159/000015408
7. Cmarik JL, Min H, Hegamyer G, et al. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci U S A. 1999;96(24):14037–14042. doi:10.1073/pnas.96.24.14037
8. Waters LC, Veverka V, Bohm M, et al. Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A. Oncogene. 2007;26(34):4941–4950. doi:10.1038/sj.onc.1210305
9. Lankat-Buttgereit B, Gregel C, Knolle A, Hasilik A, Arnold R, Goke R. Pdcd 4 inhibits growth of tumor cells by suppression of carbonic anhydrase type II. Mol Cell Endocrinol. 2004;214(1–2):149–153. doi:10.1016/j.mce.2003.10.058
10. Li Y, Wang X, Wang X, et al. PDCD4 suppresses proliferation, migration, and invasion of endometrial cells by inhibiting autophagy and NF-kappaB/MMP2/MMP9 signal pathway. Biol Reprod. 2018;99(2):360–372. doi:10.1093/biolre/ioy052
11. Yang YL, Liu P, Li D, Yang Q, Li B, Jiang XJ. Stat-3 signaling promotes cell proliferation and metastasis of gastric cancer through PDCD4 downregulation. Kaohsiung J Med Sci. 2020;36(4):244–249. doi:10.1002/kjm2.12159
12. Takaki S, Eto K. Cytoplasmic localization of programmed cell death 4 contributes to its anti-apoptotic function. Mol Cell Biochem. 2018;448(1–2):155–164. doi:10.1007/s11010-018-3322-z
13. Ma G, Guo KJ, Zhang H, et al. [Expression of programmed cell death 4 and its clinicopathological significance in human pancreatic cancer]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2005;27(5):597–600. Chinese.
14. Mudduluru G, Medved F, Grobholz R, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer-Am Cancer Soc. 2007;110(8):1697–1707.
15. Walter BA, Gomez-Macias G, Valera VA, Sobel M, Merino MJ. miR-21 expression in pregnancy-associated breast cancer: a possible marker of poor prognosis. J Cancer. 2011;2:67–75. doi:10.7150/jca.2.67
16. Zhang H, Ozaki I, Mizuta T, et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25(45):6101–6112. doi:10.1038/sj.onc.1209634
17. Li J, Fu H, Xu C, et al. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. Bmc Cancer. 2010;10:354. doi:10.1186/1471-2407-10-354
18. Wang L, Chen R, Zhang Y. miR-296-3p targets APEX1 to suppress cell migration and invasion of non-small-cell lung cancer. Oncol Lett. 2019;18(3):2612–2618. doi:10.3892/ol.2019.10572
19. Bavelloni A, Ramazzotti G, Poli A, et al. MiRNA-210: a current overview. Anticancer Res. 2017;37(12):6511–6521. doi:10.21873/anticanres.12107
20. Ding L, Gu H, Xiong X, et al. MicroRNAs involved in carcinogenesis, prognosis, therapeutic resistance and applications in human triple-negative breast cancer. Cells-Basel. 2019;8(12):1492.
21. Pfeffer SR, Yang CH, Pfeffer LM. The role of miR-21 in cancer. Drug Dev Res. 2015;76(6):270–277. doi:10.1002/ddr.21257
22. Xue X, Liu Y, Wang Y, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7(51):84508–84519. doi:10.18632/oncotarget.13022
23. Luo LJ, Wang DD, Wang J, Yang F, Tang JH. Diverse roles of miR-335 in development and progression of cancers. Tumour Biol. 2016;37(12):15399–15410. doi:10.1007/s13277-016-5385-3
24. Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99(3):671–678. doi:10.1002/jcb.20982
25. Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47(4):1223–1232. doi:10.1002/hep.22158
26. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–1033. doi:10.1074/jbc.M707224200
27. Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi:10.1038/sj.onc.1210856
28. Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13(6):580–590. doi:10.1093/neuonc/nor033
29. Qiu X, Dong S, Qiao F, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32(27):3296–3305. doi:10.1038/onc.2013.150
30. Wang M, Wang Y, Zang W, et al. Downregulation of microRNA-182 inhibits cell growth and invasion by targeting programmed cell death 4 in human lung adenocarcinoma cells. Tumour Biol. 2014;35(1):39–46. doi:10.1007/s13277-013-1004-8
31. Wang YQ, Guo RD, Guo RM, Sheng W, Yin LR. MicroRNA-182 promotes cell growth, invasion, and chemoresistance by targeting programmed cell death 4 (PDCD4) in human ovarian carcinomas. J Cell Biochem. 2013;114(7):1464–1473. doi:10.1002/jcb.24488
32. Fassan M, Pizzi M, Battaglia G, et al. Programmed cell death 4 (PDCD4) expression during multistep Barrett’s carcinogenesis. J Clin Pathol. 2010;63(8):692–696. doi:10.1136/jcp.2010.078253
33. Gao F, Wang X, Zhu F, et al. PDCD4 gene silencing in gliomas is associated with 5ʹCpG island methylation and unfavourable prognosis. J Cell Mol Med. 2009;13(10):4257–4267. doi:10.1111/j.1582-4934.2008.00497.x
34. Qi C, Shao Y, Li N, Zhang C, Zhao M, Gao F. Prognostic significance of PDCD4 expression in human salivary adenoid cystic carcinoma. Med Oncol. 2013;30(1):491. doi:10.1007/s12032-013-0491-1
35. Wang Q, Yang HS. The role of Pdcd4 in tumour suppression and protein translation. Biol Cell. 2018;110(8):169–177. doi:10.1111/boc.201800014
36. Afify M, Kamel RR, Elhosary YA, Hegazy AE, Fahim HH, Ezzat WM. The possible role of Cytochrome c and programmed cell death protein 4 (PDCD4) on pathogenesis of hepatocellular carcinoma. J Genet Eng Biotechnol. 2015;13(2):157–163. doi:10.1016/j.jgeb.2015.10.002
37. Li X, Xin S, Yang D, et al. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. J Cancer Res Clin Oncol. 2012;138:529–535. doi:10.1007/s00432-011-1121-y
38. Nong YX, Huang JL, Huang ZS, et al. Characteristics and experimental applications of human hepatocellular carcinoma cell lines. World Chin J Digestol. 2017;25:159–165. doi:10.11569/wcjd.v25.i2.159
39. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974. doi:10.1038/onc.2008.274
40. Wang J, Wang X, Li Z, Liu H, Teng Y. MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J. 2014;281(5):1355–1365. doi:10.1111/febs.12659
41. Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi:10.1126/science.1141229
42. Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31(5):766–776. doi:10.1093/carcin/bgp250
43. Zheng F, Liao YJ, Cai MY, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61(2):278–289. doi:10.1136/gut.2011.239145
44. Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106(6):1814–1819. doi:10.1073/pnas.0808263106
45. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–533. doi:10.1038/nature08199
46. Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. Bmc Cancer. 2012;12:227. doi:10.1186/1471-2407-12-227
47. Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283(19):13205–13215. doi:10.1074/jbc.M707629200
48. Wu Y, Zhu X, Shen R, Huang J, Xu X, He S. miR-182 contributes to cell adhesion-mediated drug resistance in multiple myeloma via targeting PDCD4. Pathol Res Pract. 2019;215(11):152603. doi:10.1016/j.prp.2019.152603
49. Ning FL, Wang F, Li ML, Yu ZS, Hao YZ, Chen SS. MicroRNA-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting PDCD4. Diagn Pathol. 2014;9:143. doi:10.1186/1746-1596-9-143
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.