Back to Journals » Infection and Drug Resistance » Volume 16
Microbiological Characterization and Clinical Facets of Elizabethkingia Bloodstream Infections in a Tertiary Care Hospital of Eastern India
Authors Sarathi S , Behera B , Mahapatra A, Mohapatra S , Jena J, Nayak S
Received 3 March 2023
Accepted for publication 17 May 2023
Published 24 May 2023 Volume 2023:16 Pages 3257—3267
DOI https://doi.org/10.2147/IDR.S409121
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Sushree Sarathi,1 Bijayini Behera,1 Ashoka Mahapatra,1 Sarita Mohapatra,2 Jayanti Jena,1 Saurav Nayak3
1Department of Microbiology, All India Institute of Medical Sciences [AIIMS], Bhubaneswar, Odisha, 751019, India; 2Department of Microbiology, All India Institute of Medical Sciences [AIIMS], New Delhi, 110608, India; 3Department of Biochemistry, All India Institute of Medical Sciences [AIIMS], Bhubaneswar, Odisha, 751019, India
Correspondence: Ashoka Mahapatra, Department of Microbiology, All India Institute of Medical Sciences [AIIMS], Bhubaneswar, Odisha, 751019, India, Tel +919437302030, Email [email protected]
Purpose: Elizabethkingia is an emerging non-fermenting Gram-negative bacillus (NFGNB) causing bloodstream infections (BSI) associated with high mortality. It demonstrates a unique antimicrobial profile in showing susceptibility to antimicrobials effective against Gram-positive bacteria. This study was undertaken to determine the overall frequency of Elizabethkingia BSI, associated risk factors, microbiological susceptibility, and clonal relationship of Elizabethkingia isolates using Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR).
Patients and Methods: Elizabethkingia isolates obtained from the blood culture of admitted patients (August 2020–December 2021) were identified by the VITEK 2 system and subjected to an antimicrobial susceptibility test by standard procedures. Demographics, co-morbidities, risk factors for survival, and outcome were summarized and analyzed by Chi-square test, Kaplan–Meier curve, and Cox regression. Clonal relatedness between Elizabethkingia isolates was analyzed using ERIC‑PCR fingerprinting with the “PAST: Paleontological statistics software package”.
Results: Of 13,747 blood samples received during the study period, 13.59% were culture positive, and 14.60% were NFGNBs. The frequency of Elizabethkingia spp. among all NFGNBs in BSI was 29.30%, and the overall prevalence in BSI was 4.21%. In patients with Elizabethkingia BSI, Foley’s catheter was present in 81.25% of the cases. 100% susceptibility was observed to linezolid, followed by vancomycin (98.75%) and chloramphenicol (89.5%). The 30-day mortality rate in the patients of Elizabethkingia BSI was 26.25%. The Presence of COVID-19, pneumonia, diabetes mellitus (DM), mechanical ventilation (MV), and prior antibiotics were significantly different (p< 0.05) between the survival and death groups. ERIC-PCR profile dendrogram of Elizabethkingia isolates showed ten major clusters indicating high genetic diversity.
Conclusion: Elizabethkingia was responsible for one-third of NFGNB BSI in a single-center study, with approximately 26% of 30-day all-cause mortality. Most isolates were susceptible to linezolid, vancomycin, and chloramphenicol. COVID-19 was the most significant risk factor associated with mortality. ERIC-PCR of Elizabethkingia isolates exhibited high genetic diversity.
Keywords: Elizabethkingia, bloodstream infection, risk factor, ERIC PCR typing
Introduction
Elizabethkingia, a ubiquitous non-fermenting Gram-negative bacillus (NFGNB), is gradually emerging as a significant pathogen in BSI, in addition to the traditional NFGNBs, eg, Acinetobacter, and Pseudomonas.1
With conventional microbiological techniques, the genus and species level identification of Elizabethkingia is difficult. To date, six species of Elizabethkingia have been identified- E. meningoseptica, E. miricola, E. anophelis, E. bruuniana, E. ursingii, and E. occulta. E. anophelis/E. meningoseptica are the species most associated with BSI.2–5 They have intrinsic resistance to multiple classes of antibiotics, including colistin, thus leaving few therapeutic options.6,7 Over usage of colistin has been hypothesized to be one of the reasons behind the emergence of this bacteria. Moreover, Elizabethkingia has the unique ability to acquire multi-drug resistance and survive in disinfectants enabling transmission between patients through human/inanimate reservoirs materials in the hospital environment.8 Bacterial strain typing is vital for determining the source of infection and establishment of its transmission dynamics in hospital settings. Considerable variation exists in local epidemiology, antibiotic resistance, therapeutic approach and outcome of BSI due to Elizabethkingia spp. A high rate of initial inappropriate therapy or less effective therapy may lead to poor clinical outcomes. Most of the earlier literature on NFGNB BSI had focused on Acinetobacter baumannii and Pseudomonas aeruginosa. Other NFGNBs are relatively less studied in India and abroad. Indian studies have not used molecular typing for NFGNB other than A. baumannii and P. aeruginosa. Hence, the present study was designed to determine the overall frequency, risk factors, association with 30-day mortality, and microbiological susceptibility of Elizabethkingia BSI, further to look for the clonal relationship of the Elizabethkingia isolates using Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR).
Materials and Methods
Study Setting and Sample Collection and Identification
The study was conducted at the Department of Microbiology of All India Institute of Medical Sciences, a tertiary healthcare center in Bhubaneswar, Odisha, India, from August 2020 to December 2021. All non-duplicate consecutive positive blood cultures performed in continuously monitoring blood culture/manual systems (showing GNB on Gram stain) were enrolled in the study. Positive blood culture bottles yielding pure Gram-positive cocci/bacilli/mixed flora on Gram stain were excluded.
Positively flagged bottles showing GNB on Gram stain were subcultured on 5% sheep Blood agar and Mac-Conkey agar (Hi-Media), incubated overnight at 37°C aerobically and were examined the next day. Non-lactose fermenting translucent colonies showing oxidase-positive, non-fermentative O-F-test, K/K on TSI agar and resistance to colistin were suspected to be Elizabethkingia and were subjected to provisional identification by VITEK 2 compact system GN card (BioMérieux).9 Isolates provisionally identified as Elizabethkingia by VITEK 2 system were sent for final species identification by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) [BioMérieux] at AIIMS, New Delhi.
Antibacterial susceptibility test (AST) was performed by the Kirby Bauer disc diffusion (DD) method. The recommended antimicrobial discs and the zone of inhibition for Acinetobacter baumannii were taken as interpretative criteria as there are no standard guidelines for reporting AST for Elizabethkingia in the Clinical and Laboratory Standards Institute (CLSI) M100, 2020. As per available literature, susceptibility testing of vancomycin, linezolid, and chloramphenicol was also performed by the DD method and interpreted according to CLSI standards for Enterococcus species.10
Demographic and Clinical Characteristics of the Patients
Patient-related demographic details, underlying co-morbidity, risk factors (indwelling devices, prior antibiotic usage, etc.), and outcome (discharge, ICU stay, 30-day mortality) were collected from clinical records. Prior antibiotic use was defined as any antibiotic treatment for >24 h within 1 month before the episode of infection.11 Thirty-day mortality was defined as the time from the diagnosis of Elizabethkingia bacteremia to in-hospital death within 30 days.12
ERIC PCR Analysis
DNA extraction from pure colonies was carried out using a Spin Column Qiagen miniprep kit. ERIC-PCR was performed using the primers ERIC-IR:5’-ATGTAAGCTCCTGGGGAATCAC-3’ (F) and ERIC-2:5’-AAGTAAGTGACTGGGGTGAGCG-3’ (R).13 The PCR amplicons (5μL with loading dye) were subjected to gel run using 1.5% agarose gel electrophoresis and visualized by ultraviolet transilluminator (Syngene, Germany). Clonal relatedness between the isolates of Elizabethkingia was analyzed using ERIC PCR fingerprinting with the Paleontological Statistics (PAST) software package. Normalization steps were included to analyze the DNA polymorphism patterns produced by ERIC PCR fingerprinting to ensure an adequate gel-to-gel banding pattern comparison. Band scoring was used to identify bands in each lane and to make the fingerprint. Dendrograms were generated for the ERIC PCR gels using the Dice similarity coefficient and interpreted using arithmetic averages with 1% optimization and 1% position tolerance. Elizabethkingia isolates with a similarity exceeding 60% were considered to be clonally related.13
Statistical Analysis
Continuous variables were described as mean, median (Student’s t-test), and categorical variables as frequency counts/percentages (Chi-square test/Fisher’s exact test). The risk factors were estimated using the Chi-square test and Kaplan–Meier survival analysis to screen possible factors related to 30-day mortality or survival. Statistically significant variables (p< 0.10) in the univariate analyses were used to construct the multivariate model (Cox regression analysis). A two-sided p-value < 0.05 was considered to be statistically significant.
Results
A total of 13,747 blood culture samples were processed during the study period, of which 13.59% (1869/13747) were culture positive and 14.60% (273/1869) were NFGNB. The frequency of Elizabethkingia spp. among all NFGNB in BSI was 29.30% (80/273) and the overall prevalence in BSI was 4.21% (80/1896). All 80 Elizabethkingia isolates were identified as E. meningoseptica in VITEK-2 compact system; however, in MALDI-TOF MS, all were identified as E. anophelis.
Demographic Details, Risk Factor Analysis, and 30-Day Mortality
The mean age of patients who developed Elizabethkingia BSI was 47 years; maximum isolates were obtained from the age group 60–69 years – 20.00% (16/80) and more from males (61.25%, 49/80) as compared to females (38.75%, 31/80). Elizabethkingia isolates were equally obtained from ICU & non-ICU settings (40/80, 50% each). Among the ICUs, the majority were from COVID ICU (16/40, 40%) and among wards, the majority were from Medicine wards (12/40, 30%). The 30-day mortality rate in the patients of Elizabethkingia BSI was 26.25% (21/80).
The risk factors of Elizabethkingia BSI are listed in Table 1. Among the risk factors, COVID-19, pneumonia, prior antibiotic intake, MV & DM differed significantly between the survival and death groups. According to the Kaplan–Meier survival analysis for univariate analysis (Figure 1), pneumonia was significantly associated with the survival rate. Further, multivariate Cox regression analysis showed that those having pneumonia had 7.7 times more risk of death within 30 days of culture positivity (Table 2).
 |
Table 1 Risk Factors Related to 30-Day Mortality of Elizabethkingia BSI |
 |
Table 2 Multivariate Cox Regression Model Analysis |
 |
Figure 1 Kaplan–Meier curve comparing the survival rate between patients with and without pneumonia. |
Antibacterial Susceptibility Results
Susceptibility to linezolid was 100%, vancomycin 98.75%, and chloramphenicol 89.5%. All the isolates of Elizabethkingia were MDR, and more than 75% were resistant to carbapenems, aminoglycosides, and third- and fourth-generation cephalosporins (Table 3).
 |
Table 3 Susceptibility Pattern of 80 Tested Elizabethkingia Isolates (n=80) |
ERIC PCR Typing Results
The timeline for the isolation of Elizabethkingia spp. from BSI has been depicted in Figure 2. From the fingerprints obtained from ERIC typing (Figure 3), 10 major clusters were identified (Figure 4). Among those, cluster A had the majority of the isolates (35 strains), and 10 out of them were retrieved from COVID ICU intermittently throughout the year from August 6, 2020 to August 1, 2021, 8 were from Central ICU (CICU) [from September 9, 2020 to September 21, 2021]. The majority of the isolates analyzed in our study were genetically diverse. Few strains have similar ERIC profiles.
 |
Figure 2 Timeline for isolates of Elizabethkingia spp. from BSI. |
 |
Figure 3 ERIC-PCR generated DNA fingerprints of Elizabethkingia isolates. Notes: Marker: 100bp DNA ladder, Lanes 1–12: Elizabethkingia isolates. |
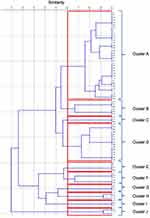 |
Figure 4 Phylogenetic relationships among the Elizabethkingia BSI isolates derived from analysis of ERIC PCR profiles. |
Discussion
Since the last decade, NFGNBs other than P. aeruginosa and A. baumannii have emerged as pathogens responsible for BSI in healthcare settings. Previously, these bacteria isolated from clinical samples were regarded as colonizers rather than pathogens. In the present cross-sectional laboratory-based study, 80 Elizabethkingia BSI cases were reviewed from August 2020 to December 2021 in a tertiary care hospital in Eastern Odisha. The prevalence of NFGNB in BSI during the study period was 14.39% (273/1896). Sarwat et al reported the prevalence of NFGNB to be 16.6% (176/1041) from a tertiary care center in Uttar Pradesh, India.14 Gautam et al, in a study from Chandigarh, India, had reported a low prevalence of NFGNB (0.9%, 113/12331) in BSI.15 The institutional variation of NFGNB prevalence in BSI may be due to different patient profiles and laboratory practices.
The frequency of Elizabethkingia spp. among all NFGNB BSI in our study was 29.30% (80/273) and the overall prevalence of Elizabethkingia spp. among all BSI was 4.21% (80/1896). In a study by Singh et al. Elizabethkingia spp. were mostly isolated from blood samples (52.6%, 10/19) from an ICU in North India.16
As highlighted by previous studies, the species-level identification of Elizabethkingia by commercially available identification systems such as VITEK 2 remains suboptimal. In our study, all the isolates (80) of Elizabethkingia meningoseptica identified by the VITEK 2 compact system were confirmed to be E. anophelis by MALDI-TOF MS (BioMérieux). E. anophelis is often misidentified as E. meningoseptica and remains underreported in most clinical studies.16 In a multicentre study from Taiwan, they could discriminate three major species of Elizabethkingia using the amended database of the MALDI-TOF VITEK MS system.6 Elizabethkingia anophelis was first discovered in the gut of the Anopheles mosquito; hence, its name is anophelis. However, it was later shown that mosquitoes were unlikely to be the vector of transmission in E. anophelis-associated infections. Further research and work postulated that contaminated hospital environments, infected catheters, and intravenous infusions and fluids acted as reservoirs for E. anophelis.17 As per a study from Taiwan, 92 isolates of Elizabethkingia spp. (E. menigoseptica – 20, E. anophelis – 72) were identified from various clinical samples between January 2005 and June 2018. They observed that the frequency of isolation of E. meningoseptica (2/20) from CSF was significantly (p 0.045) more as compared to E. anophelis (0/72). When the susceptibility of each species was compared, E. meningoseptica exhibited significantly lower susceptibility rates to piperacillin-tazobactam (p = 0.02), minocycline (p < 0.001), and levofloxacin (p = 0.025) and had a higher rate of a target gene mutation for fluoroquinolone as compared to E. anophelis.7 Because of this, empirical antimicrobial therapy of E. meningoseptica should be more rigorous.
Species-level identification of Elizabethkingia by traditional methods is a challenge. For precise identification of species, tools such as MALDI-TOF MS and whole-genome sequencing are required, which are not available in most hospital laboratories. Thus, difficulties in species-level identification of Elizabethkingia by traditional methods have hampered the understanding of the spectrum of infections by this genus in humans. Moreover, the misidentification of Elizabethkingia spp. could underestimate the number of cases attributed to the bacterium. In Southeast Asian countries, E. meningoseptica has usually been ascribed to hospital outbreaks, whereas E. anophelis was reported to be the main Elizabethkingia spp. associated with hospital settings in Singapore.18
The mean age of patients with E. anophelis BSI in our study was 47 years, with the majority being male (61.25%). The demographic profile of the patients enrolled in our study matches the report by Ghafur et al where the mean age of the patients developing Elizabethkingia BSI was 48.4 years. In their study population of 11 immunocompromised patients, seven were males (63.63%), and four were females (36.36%).19 Elizabethkingia isolates were equally obtained from patients admitted to ICU & non-ICU (40/80, 50% each) of our hospital. Among the ICUs, the majority were from COVID ICU (16/40 40%) and among wards, the majority were from Medicine wards (12/40 30%).
In the current study, most patients developing Elizabethkingia BSI (65/80, 81.25%) had more than one underlying disease. The most frequently encountered diseases leading to BSI were pneumonia (COVID & non-COVID) (29/80, 36.25%) followed by COVID-19 (COVID-19 positive status) (27/80, 33.75%) and malignancies (6/80, 7.5%). The most common underlying co-morbidities associated with Elizabethkingia BSI were DM (20/80, 25%), hypertension (14/80, 17.5%), cardiovascular diseases (12/80, 15%), diseases involving the central nervous system (7/80, 8.75%) and digestive tract (12/80, 15%). In a similar study by Seong et al from South Korea, most subjects who contracted Elizabethkingia infection had pneumonia (n = 186) and malignancy (n = 100) as the most common underlying disease among the 210-study population. The non-survivors presented a higher percentage with regard to previous ICU admission [40 (75.5%) vs 89 (56.7%), p = 0.015], central venous catheter use [48 (90.6%) vs 121 (77.1%), p = 0.032], and immunocompromised status [24 (45.3%) vs 42 (26.8%), p = 0.012].20
In the present study, the all-cause mortality (within 30 days) from BSI caused by NFGNB was 27.67% (57/206), and mortality due to Elizabethkingia spp. alone was 26.25% (21/80). This was in agreement with the studies from Hong Kong and Korea, where the overall 30-day mortality due to bacteremia and pneumonia was 23.5% and 25.2% (53/210), respectively.17,21 Univariate analysis of risk factors showed that the odds of individuals having pneumonia, COVID-19, DM, MV, and prior antibiotic intake were higher in the mortality group with Elizabethkingia BSI than in the survival group. Those having pneumonia had 7.7 times more risk of death within 30 days of culture positivity (Table 2).
However, a high mortality rate, almost twice (41.6%,10/24) of the aforementioned studies, has been reported from China.22 They have justified inadequate empirical antimicrobial therapy as an independent risk factor for mortality by multivariate analysis model. Although the exact cause for the mortality observed in our study could not be explained, it is assumed that the coexistence of COVID-19 may be a significant contributing factor. BSI pathogens and resistance profiles differ in patients with COVID-19. Moreover, severe COVID-19 is associated with increased pro-inflammatory markers leading to increased susceptibility to bacterial and fungal infections.23
Elizabethkingia isolates showed very good in-vitro susceptibility to linezolid (80/80, 100%), vancomycin (79/80, 98.75%), chloramphenicol (73/80, 89.5%), ciprofloxacin (70/80, 87.50%), cefoperazone-sulbactam (63/80, 78.75%) and cotrimoxazole (60/80, 75%). Poor susceptibility was noted for gentamicin (52/80, 65.00%), amikacin (45/80, 56.25%), cefepime (9/80, 11.25%), imipenem (4/80, 5.00%), meropenem (3/80,3.75%), ceftriaxone (3/80, 3.75%), ampicillin (2/80, 2.50%) and ceftazidime (2/80, 2.50%). Several countries have reported the antimicrobial susceptibility of E. anophelis to exhibit resistance to most of the β-lactams, β-lactam combination agents, aminoglycosides, quinolones, and carbapenems irrespective of the testing techniques – Hong Kong (21/21, 100%), China (100% MDR and 20 were extensively drug-resistant, n=197), Taiwan (ciprofloxacin – 89%, cotrimoxazole – 96%) and Lucknow, India (100% resistance to levofloxacin and ciprofloxacin).6,16,17,24
The widespread resistance to various β-lactams could be due to the production of metallo-β-lactamases (coded by Bla B and Bla (GOB) genes).25 Few studies have reported the correlation between biofilm formation and antibiotic resistance in Elizabethkingia species from humans.26,27 Singh et al from India reported 100% susceptibility to minocycline and piperacillin-tazobactam, but the susceptibility to vancomycin was very poor (32.6%).16 Inconsistent AST results were reported for some antibiotics, possibly due to insufficient sample size and/or origin of the strains from different geographic regions. Hence, its performance in comparison to the recommended method (broth/agar dilution) for non-Enterobacterales needs to be evaluated. However, the advantage of linezolid and vancomycin therapy over conventional Gram-negative agents for Elizabethkingia BSI must be explored.
In our study, 10 unique clusters of ERIC were observed among the 80 Elizabethkingia isolates exhibiting genetic heterogeneity. Before 2020, we had sporadic isolation of Elizabethkingia from BSI cases. Over the study period, there was a continuous common source type of clustering of isolates during the initial 6 months, followed by a decline, which peaked in the later months, followed by a sharp decline (Figure 2). However, extensive environmental investigation of all water sources, moist environmental sources, IV fluids, and disinfectants could not point toward any source. However, some isolates had similar ERIC profiles and showed a genetically homogeneous pattern, but no correlation was found between the clonally similar strains and their antibiotic susceptibility pattern. Only a few isolates showed a conserved banding profile based on ERIC analyses, which indicated the rapid dissemination of similar clonal groups. Three unique clusters of ERIC in 13 isolates of E. meningoseptica were reported from an outbreak in the neonatal and pediatric unit of a hospital in Turkey by Ceyhan et al. As per their report, all the three clusters were caused by an epidemiologically related strain, and the index case identified from the NICU was assigned to be temporarily responsible for the clonal outbreak.28
The occurrence of a few clonally similar groups among our isolates could be due to the spread of antibiotic resistance through the dissemination of different β-lactamases or clonal transmission. However, the lack of association of these clonally similar isolates with their respective antibiotic sensitivity pattern in our study rules out the above possibility. Antibiotic selection pressure could be one of the significant reasons which need to be validated further.
PFGE is known as the “gold standard” among all the commonly used molecular typing methods worldwide. However, it has some drawbacks, like the requirement of comparatively large DNA quantities, high-cost specific equipment, and is time-consuming. On the other hand, the ERIC-PCR method is sensitive, rapid, low cost, repeatable, resolving, reliable, and easy to handle. Therefore, this typing method has been broadly applied to the genotypic typing of microbes.29
Strengths of This Study
NFGNBs, except A. baumannii and P. aeruginosa in BSIs, are less studied in India and abroad. To our knowledge, this is the first study from Eastern India (Odisha) to look for their prevalence, risk-factor association and clonal relationship using ERIC-PCR.
Limitations in Our Study
The sample size of the present study was small. A multicentric research with a larger sample size may help better establish the genetic diversity among the Elizabethkingia BSI isolates from this region. Moreover, since there are no interpretative standards for Elizabethkingia spp., AST with DD method was adopted, which may not be reliable in determining susceptibility and guiding therapy.
Conclusion
The overall prevalence of Elizabethkingia BSI in our study was 4.21%, with a 30-day mortality rate of 26.25%. Significant risk factors were COVID-19, pneumonia, prior antibiotic intake, mechanical ventilation and DM. For the speciation of Elizabethkingia isolates, MALDI-TOF MS has shown promising results. High in vitro susceptibility was shown to linezolid, vancomycin, and chloramphenicol, and there was a continuous common source type of clustering of isolates during the initial 6 months. Thus, healthcare professionals (HCPs) need to heighten awareness about Elizabethkingia as an emerging pathogen for BSI among hospitalized patients. Stringent infection control measures of the hospital environment and hand hygiene compliance of HCPs are necessary to prevent the potential threat of Elizabethkingia outbreaks in ICUs.
Ethics Approval and Informed Consent
We conducted the study in accordance with the amended Declaration of Helsinki. Written informed consent was obtained from all patients. The study was approved by the institutional ethics committee of All India Institute of Medical Sciences [AIIMS], Bhubaneswar, Odisha (Ref Number: IEC/AIIMS BBSR/PG Thesis/2020-21/59 dated 8 July 2020).
Consent for Publication
All details of any content and images can be published, and every author has shown the article content to be published. Patient agreed to publish the details of the case and signed the written informed consent for the case details to be published.
Acknowledgments
The authors thank Mrs Alakanada Mohapatra for her technical support.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Choi MH, Kim M, Jeong SJ, et al. Risk factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg Infect Dis. 2019;25(1):42–51. doi:10.3201/eid2501.171985
2. Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. Int J Syst Evol Microbiol. 2005;55(3):1287–1293. doi:10.1099/ijs.0.63541-0
3. Li Y, Kawamura Y, Fujiwara N, et al. Chryseobacterium miricola sp. nov., A novel species isolated from condensation water of space station mir. Syst Appl Microbiol. 2003;26(4):523–528. doi:10.1078/072320203770865828
4. Kämpfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61(11):2670–2675. doi:10.1099/ijs.0.026393-0
5. Nicholson AC, Gulvik CA, Whitney AM, et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie van Leeuwenhoek. 2018;111(1):55–72. doi:10.1007/s10482-017-0926-3
6. Cheng YH, Perng CL, Jian MJ, et al. Multicentre study evaluating matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically isolated Elizabethkingia species and analysis of antimicrobial susceptibility. Clin Microbiol Infect. 2019;25(3):340–345. doi:10.1016/j.cmi.2018.04.015
7. Lin JN, Lai CH, Yang CH, Huang YH. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between elizabethkingia meningoseptica and elizabethkingia anophelis isolated in Taiwan. JCM. 2018;7(12):538. doi:10.3390/jcm7120538
8. Lin JN, Lai CH, Yang CH, Huang YH. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms. 2019;7(9):295. doi:10.3390/microorganisms7090295
9. Chaudhury N, Paul R, Misra RN, Chaudhuri SS, Mirza S, Sen S. Evaluating the trends of bloodstream infections by nonfermenting gram negative bacilli among the patients in a tertiary care hospital of western part of India and its antibiogram. IntJCurrMicrobiolAppSci. 2019;8(01):1149–1162. doi:10.20546/ijcmas.2019.801.121
10. Chiu CT, Lai CH, Huang YH, Yang CH, Lin JN. Comparative analysis of gradient diffusion and disk diffusion with agar dilution for susceptibility testing of Elizabethkingia anophelis. Antibiotics. 2021;10(4):450. doi:10.3390/antibiotics10040450
11. Lai CH, Wong WW, Chin C, et al. Central venous catheter-related Stenotrophomonas maltophilia bacteraemia and associated relapsing bacteraemia in haematology and oncology patients. Clin Microbiol Infect. 2006;12(10):986–991. doi:10.1111/j.1469-0691.2006.01511.x
12. Hashimoto T, Komiya K, Fujita N, et al. Risk factors for 30-day mortality among patients with Stenotrophomonas maltophilia bacteraemia. Infect Dis. 2020;52(6):440–442. doi:10.1080/23744235.2020.1734653
13. Asgarani E, Ghashghaei T, Soudi MR, Alimadadi N. Enterobacterial repetitive intergenic consensus (ERIC) PCR based genetic diversity of Xanthomonas spp. and its relation to xanthan production. Iran J Microbiol. 2015;7(1):38–44.
14. Sarwat T, Yousuf M, Khan AS, Kakru DK, Dutta R. Prevalence and antibiogram of non-fermenting Gram-negative bacilli in blood stream infections: study in a tertiary care centre, Western Uttar Pradesh, India. Trop Doct. 2021;51(3):322–325. doi:10.1177/0049475520979298
15. Gautam V, Thapar R, Ray P, Samanta P. Emerging resistance of non-fermenting gram negative bacilli in a tertiary care centre. Indian J Pathol Microbiol. 2011;54(3):666. doi:10.4103/0377-4929.85150
16. Singh S, Sahu C, Singh Patel S, Singh S, Ghoshal U. Clinical profile, susceptibility patterns, speciation and follow up of infections by Elizabethkingia species: study on a rare nosocomial pathogen from an intensive care unit of north India. New Microbes New Infect. 2020;38:100798. doi:10.1016/j.nmni.2020.100798
17. Lau SKP, Chow W-N, Foo C-H. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep. 2016;10:26045. doi:10.1038/srep26045
18. Zajmi A, Teo J, Yeo CC. Epidemiology and characteristics of Elizabethkingia spp. infections in Southeast Asia. Microorganisms. 2022;10(5):882. doi:10.3390/microorganisms10050882
19. Ghafur A, Vidyalakshmi PR, Priyadarshini K, Easow JM, Raj R, Raja T. Elizabethkingia meningoseptica bacteremia in immunocompromised hosts: the first case series from India. South Asian J Cancer. 2013;02(04):211–215. doi:10.4103/2278-330X.119912
20. Seong H, Kim JH, Kim JH, et al. Risk factors for mortality in patients with elizabethkingia infection and the clinical impact of the antimicrobial susceptibility patterns of Elizabethkingia species. JCM. 2020;9(5):1431. doi:10.3390/jcm9051431
21. Wan Q, Ye Q, Huang F. The bacteremia caused by non-lactose fermenting gram-negative bacilli in solid organ transplant recipients. Surg Infect. 2015;16(5):479–489. doi:10.1089/sur.2015.005
22. Li Y, Liu T, Shi C, et al. Epidemiological, clinical, and laboratory features of patients infected with Elizabethkingia meningoseptica at a tertiary hospital in Hefei City, China. Front Public Health. 2022;10:964046. PMID: 36225778; PMCID: PMC9549487. doi:10.3389/fpubh.2022.964046
23. Bhatt K, Agolli A, Patel MH, et al. Guerra Del Castillo R, Sanchez-Gonzalez M. High mortality co-infections of COVID-19 patients: mucormycosis and other fungal infections. Discoveries. 2021;9(1):e126. PMID: 34036149; PMCID: PMC8137279. doi:10.15190/d.2021.5
24. Hu S, Lv Y, Xu H, Zheng B, Xiao Y. Biofilm formation and antibiotic sensitivity in Elizabethkingia anophelis. Front Cell Infect Microbiol. 2022;28(12):953780. doi:10.3389/fcimb.2022.953780
25. Gonzalez LJ, Vila AJ. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-β-lactamase BlaB. Antimicrob Agents Chemother. 2012;4:1686–1692. doi:10.1128/AAC.05835-11
26. Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother. 2018;73(9):2497–2502. doi:10.1093/jac/dky197
27. Tang HJ, Lin YT, Chen CC, et al. Molecular characteristics and in vitro effects of antimicrobial combinations on planktonic and biofilm forms of Elizabethkingia anophelis. J Antimicrob Chemother. 2021;76:1205–1214. doi:10.1093/jac/dkab018
28. Ceyhan M, Yıldırım I, Tekelı A, et al. A Chryseobacterium meningosepticum outbreak observed in 3 clusters involving both neonatal and non-neonatal pediatric patients. Am J Infect Control. 2008;36(6):453–457. doi:10.1016/j.ajic.2007.09.008
29. Guimarães A de S, Dorneles EMS, Andrade GI, et al. Molecular characterization of Corynebacterium pseudotuberculosis isolates using ERIC-PCR. Vet Microbiol. 2011;153(3–4):299–306. doi:10.1016/j.vetmic.201
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
