Back to Journals » Cancer Management and Research » Volume 12
MDM2-C Functions as an E3 Ubiquitin Ligase
Authors Kim JY , Lee R , Xiao G , Forbes D , Bargonetti J
Received 7 May 2020
Accepted for publication 2 August 2020
Published 24 August 2020 Volume 2020:12 Pages 7715—7724
DOI https://doi.org/10.2147/CMAR.S260943
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Jun Yeob Kim,1 Rusia Lee,1,2 Gu Xiao,1 Dominique Forbes,1 Jill Bargonetti1– 3
1The Department of Biological Sciences, Hunter College, City University of New York, New York, NY, USA; 2Biology PhD Program, The Graduate Center of Biology, City University of New York, New York, NY, USA; 3Department of Cell and Developmental Biology, Weill Cornell Medical College, New York, NY 10021, USA
Correspondence: Jill Bargonetti The Department of Biological Sciences
Hunter College, Belfer Building, 413 East 69 th Street, New York, NY 10021, USA
Tel +1 212-896-0465
Email [email protected]
Background: Mouse double minute 2 (MDM2) is an E3 ubiquitin ligase that is over-expressed in many cancers and regulates target proteins through ubiquitination. Full-length MDM2 (MDM2-FL) is best known for targeting wild-type p53 for degradation by the proteasome, but the functions of the many splice variants of MDM2 are under-explored. The three well-studied alternative MDM2 isoforms are MDM2-A/ALT2, MDM2-B/ALT1, and MDM2-C/ALT3. MDM2-A and MDM2-B are capable of down-regulating MDM2-FL activity and have transforming activity in cancers with mutant p53. The MDM2 isoform MDM2-C is over-expressed in breast cancer and correlates with decreased survival in the context of mutant p53 expression. Therefore, MDM2-C requires further study to determine if it has biochemical activities similar to MDM2-FL. Hypothesis: We hypothesized that like MDM2-FL, the MDM2-C isoform (lacking exons 5– 9 and containing a full C-terminal RING finger sequence) would maintain E3 ubiquitin ligase activity.
Materials and Methods: In order to explore the biochemical function of MDM2-C, we used an in vitro ubiquitination assay and a glutaraldehyde cross-linking assay.
Results: Here we report, for the first time, that MDM2-C has E3 auto-ubiquitin ligase activity, which can promote ubiquitination of wild-type p53 and mutant p53 R273H, and also can form a protein–protein interaction with p53 proteins.
Conclusion: This information strongly positions MDM2-C as a protein with biochemical activities that may explain the varied outcomes observed in patients with high-level expression of MDM2-C in the presence of wild-type p53 versus mutant p53.
Keywords: MDM2 splice variants, mutant p53, wild-type p53, breast cancer
Introduction
The MDM2 E3 ubiquitin ligase belongs to a category of enzymes that regulate protein level by attaching ubiquitin, a small protein, to their substrates.1–3 An important example of an E3 ubiquitin ligase enzyme is MDM2, a protein composed of 491 amino acid residues that interact specifically with polypeptides and aids in the final step of attachment of ubiquitin passed along from the E1 and E2 conjugating enzymes.4 The region of MDM2 that performs the E3 ubiquitin ligase function is the really interesting new gene (RING) domain, located at the C-terminal end of the protein.5,6 The best-known ubiquitination target of MDM2 is the p53 transcription factor that can act as a tumor suppressor protein when stress signals allow for the disruption of the p53-MDM2 complex.5–9 MDM2 down-regulates p53 by binding to the protein and directing it for proteolysis by the proteasome.6,9-12 Structural analysis of MDM2 homodimers, and heterodimers with its related RING domain protein MDMX, make it clear that the C-terminus of MDM2 can offer differential regulatory activities under different circumstances.13 This suggests that alternative spliced forms of MDM2 that retain their C-termini might possess biochemical functions for cellular regulation. Stress regulates the alternative splicing of MDM2 and contributes an additional level for regulating cellular pathways.14–16 The three most well-studied splice variants of MDM2 result from exon skipping and are MDM2-A/ALT2, MDM2-B/ALT1, and MDM2-C/ALT3.17,18 MDM2-C is highly expressed in breast cancers and maintains a large amount of the original MDM2 structure, keeping a portion of the N-terminal p53 interaction domain, the central acidic domain and zinc finger domains, and the C-terminal RING domain.19,20
The MDM2-A and MDM2-B isoforms contain E3 ligase activity but the potential for the MDM2-C isoform to regulate ubiquitination pathways has not been explored.21 The regulation of MDM2 auto-ubiquitination is a central hub for regulating other pathways.4 In addition to auto-ubiquitination, MDM2 can ubiquitinate oncogenic mutant p53 although it does not actively target mutant p53 proteins for proteasome-mediated degradation.22,23 This is because the interaction between MDM2 and mtp53 results in a complex with different interactions due to mtp53 not binding site-specifically with DNA.24 We were particularly interested to determine the activity of MDM2-C for auto-ubiquitination and ubiquitination of oncogenic mutant p53 proteins. The auto-ubiquitination ability of MDM2-FL regulates its level during cell stress.6,25 MDM2-C transcript and protein are present in liposarcoma and breast cancer tissue and cell-lines and MDM2-C is known to promote tumorigenesis in cells that do not express wild-type p53.19,20 This suggests that MDM2-C might function as an E3 ubiquitin ligase for itself and other proteins and the resulting ubiquitination patterns that might promote cancer in the presence of mtp53. The ubiquitination of proteins does not always signal for increased protein degradation it can also be a post-translational modification that can alter cellular signaling.1
MDM2-C lacks exons 5–9 due to alternative splicing but the MDM2 RING domain is kept intact, as the alternative splicing does not influence exon 12.18,19 To test if MDM2-C maintained E3 ubiquitin ligase activity, we expressed the protein in bacteria, purified it, and carried out in vitro ubiquitination assays. In this way, we examined self-ubiquitination, wild-type p53 (wtp53) ubiquitination, and mutant p53 (mtp53) R273H ubiquitination. We detected, for the first time, that MDM2-C has E3 ubiquitin ligase activity. The type of ubiquitination link was not identified and the type of cellular signal evoked by MDM2-C mediated ubiquitination is an area for future study.
Materials and Methods
Plasmid Construction
pRSETA-mdm2-C was constructed by inserting mdm2-C into pRSETA by swapping out mdm2-FL from pRSETA-hdm2, a generous gift from Dr. Lindsay Mayo. Restriction enzymes EcoRI and XhoI (New England Labs) were used to excise hdm2 from pRSETA-hdm2 and to create overhangs for mdm2-C. pT7-CFE1-CGST-HA-His-mdm2-C was used to provide the mdm2-C cDNA sequence. Ligation was performed to insert Mdm2-C into pRSETA.
Protein Expression and Purification
MDM2-FL and MDM2-C were overexpressed in E. Coli (BL21DE3) with vectors pRSETA-hdm2 and pRSETA-mdm2-C, respectively. The clones were grown in LB media with 100 µM ZnCl2 and 0.5% (w/v) glucose supplements until OD600 reached early log phase (0.1–0.2). The strains were induced with 100 µM IPTG for 18 h at 15°C. Cells were harvested with centrifugation at 6000 rpm for 15 min and lysed with Buffer A (25 mM HEPES pH 7.9, 0.2% Triton-X, 5 mM DTT, 1 M KCl, 1 mM PMSF) via sonication. Proteins were bound to Ni-NTA agarose beads (Qiagen) with Buffer A, washed extensively with Buffer B (Buffer A + 10 mM imidazole), and eluted with Buffer C (Buffer A + 300 mM imidazole) for 1 h in cold temperature. The eluted proteins were dialyzed in Buffer D (50 mM HEPES pH 7.9, 100 mM NaCl, 10% (v/v) glycerol, 1 mM DTT) set in pH 7.4 overnight. For purification, Wtp53 and mtp53 were expressed in Sf-21 cells infected with human wtp53 or mtp53 recombinant baculovirus. After 48 h of infection, the cells were harvested, extracted, and immunopurified using monoclonal antibody PAb 421 cross-linked to protein A-Sepharose.
In vitro Ubiquitination Assay
The assay was carried out in 1X ubiquitination buffer (50 mM Tris pH 7.5, 1 mM DTT, 2 mM ATP, 5 mM MgCL2) with the addition of a combination of the following reaction components: 200 nM Recombinant Human His6 Ubiquitin-activating Enzyme/UBE1), 1 µM Recombinant Human UbcH5c/UBE2D3 Cat. #E2-627 (Boston Biochem), 100 µM Recombinant Human Ubiquitin Cat. #U-100H (Boston Biochem), and 1 µg of MDM2-FL or MDM2-C at 37°C for 1 h. Two micrograms of purified wild-type p53 or mutant p53 R273H was incubated in a final volume of 20 µL. Wild-type p53 and mutant p53 R273H were expressed in Sf-21 cells infected with human wtp53 or mtp53 recombinant baculovirus and purified as previously described.26 The reactions were terminated with 4X Nu-PAGE Lithium Dodecyl Sulphate and 50mM DTT and were subjected to electrophoresis on a 4–12% SDS-PAGE gradient gel.
Western Blot
Samples were boiled at 70°C for 10 minutes and added 100 mM iodoacetamide (Sigma) to prevent formation of disulfide bridges. As high as 4–12% of gradient SDS-PAGE gel (Invitrogen) was run to separate the protein samples from in vitro ubiquitination assay, and 10% SDS-PAGE gel was run to separate the protein samples for confirming purification. Polyvinylidene fluoride (GE Life Science), or PVDF, membrane was used in electrotransfer of proteins. The membrane was blocked with 5% non-fat milk in 1x PBS/0.1% Tween-20 (PBST), followed by 2x wash with 1x PBST and incubation with primary antibody overnight at 4°C. Washes were done 2x with 1X PBST, and the membrane was incubated with secondary mouse or rabbit antibody, Cy-3 and Cy-5, respectively (GE Life Science), for 1 h at room temperature in a dark setting. Further washes were done 3x with 1X PBST and 1x with 1x PBS to wash the detergent. The membrane was air-dried at 37°C for 45 minutes and proceeded for visualization with Typhoon FLA 7000 biomolecular imager (GE Life Science).
Glutaraldehyde Crosslinking Assay
The cross-linking assay was performed in 1x ubiquitination buffer using 1 µg of purified wild-type p53 and 2 µg of purified MDM2-C dialyzed in dialysis buffer. Reactions were carried out in the presence or absence of 0.005% glutaraldehyde. Samples were placed on a shaker at room temperature for 20 minutes and the reaction was stopped with the addition of 4X Nu-PAGE Lithium Dodecyl Sulphate sample buffer and 50 mM DTT. Samples were heated at 70°C for 10 minutes. Iodoacetamide at a final concentration of 100 mM was added to cooled samples, which were then run on a 4–12% SDS-PAGE gradient gel.
Antibodies
Figure legends articulate the antibodies used for the specific experiments. When the p53 mix was used to detect wtp53 and mtp53 it was composed of 3 different monoclonal supernatant p53 antibodies, 1801, 240, and 421, detecting N-terminus, Central, and C-terminus regions, respectively. We also used the primary p53 monoclonal antibody DO1 (Santa Cruz). A variety of antibodies were used to detect MDM2-FL and MDM2-C for in vitro ubiquitination assay and purification. Human/Mouse/Rat MDM2/HDM2 polyclonal antibody (R&D System), MDM2-C antibody 7C7, and MDM2-FL antibody 2A9, both of which are monoclonal supernatant MDM2 antibodies, were used for MDM2-FL and/or MDM2-C detection.
Results
Purified MDM2-FL and MDM2-C are Recognized by Distinct Monoclonal Antibodies
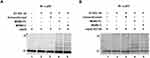
MDM2-C retains the RING domain after alternative splicing, but no studies have yet determined if MDM2-C acts as an E3 ligase. In order to test for this important biochemical activity, we bacterially expressed and purified MDM2-C and MDM2-FL (Figure 1). MDM2-C is an isoform of MDM2-FL and therefore it was critical for us to know that we were comparing the different polypeptides. MDM2-FL and MDM2-C were purified from an E. coli system as His-tagged proteins using nickel beads. Both Pierce reversible staining for membrane and Coomassie Blue for in-gel detection were used to determine the quality of purification. In Figure 1 we are making it clear that we are working with the two different versions of MDM2 (ie, MDM2-FL and MDM2-C). The membrane staining (shown in Figure 1A) was carried out to assess the amount and quality of the protein that was used for subsequent Western blot analysis. We observed a single band for MDM2-FL in lane 1 at 102 kD and MDM2-C in lane 2 at 76 kD (Figure 1A, see arrows). The purification scheme was reproducible and Coomassie Blue staining demonstrated some co-migrating protein bands from untransformed bacteria that were not reactive with any of the MDM2 antibodies described below (see Supplementary Figure S1). Untransformed purification samples were used as a negative control in biochemical assays and the Western blot indicated that there was no MDM2 present in the sample (Figure S1).
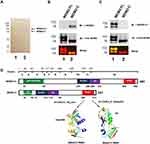 |
Figure 1 Bacterially expressed MDM2-FL and MDM2-C can be recognized by distinct monoclonal antibodies. (A) pRSETA-mdm2-C was constructed from pRSETA-hdm2,33 a generous gift from Dr. Lindsay Mayo, by PCR-mediated cloning. MDM2-FL and MDM2-C were overexpressed in BL21DE3 E. coli strains with plasmids pRSETA-hdm2 and pRSETA-mdm2-C, respectively. 2 µg of the bacterially purified MDM2-FL and MDM2-C, lanes 1 and 2, respectively, were loaded into 10% SDS-PAGE gel. Proteins were transferred to PVDF membrane. The quality of purification was confirmed by using either Pierce reversible staining for membrane or Coomassie Blue for in-gel detection. Arrows indicate the bands for MDM2-FL and MDM2-C. (B) Purified MDM2-C and (C) MDM2-FL proteins on a Western Blot visualized with either seconday anti-mouse or anti-rabbit antibody conjugated with Cy-3 and Cy-5. MDM2-FL-specific monoclonal antibody 2A9, or MDM2-C-specific monoclonal antibody 7C7 in combination with R&D polyclonal antibody (which detects both MDM2-FL and MDM2-C). The bottom image is the superimposition of bands probed with two antibodies. (D) Schematic structural domains of MDM2-FL and MDM2-C. Mdm2 gene consists of 12 exons. Known motifs are represented in color, Green: p53-binding domain; Blue: Acidic domain; Purple: zinc-finger domain; Red: RING domain (really interesting new gene). The domain interface clusters and cartoon representation of the complex of MDM2-C RING and UQ_con are show (E2 Ubiquitin-Conjugating Enzyme D2) and ubiquitin are shown. This data represents results from three independent experiments. |
We used antibody detection of the MDM2-FL and MDM2-C isoform proteins with antibodies for specific MDM2 variants to validate the production of the isoforms (Figure 1B and C). The antibodies have different recognition epitopes and therefore can be used to validate MDM2-FL or MDM2-C production but the Western blots were not used to compare the overall levels of the proteins. We used a monoclonal antibody recognizing just MDM2-C (as previously described in Loo et al20) to validate cloning and expression of the MDM2-C isoform (Figure 1B, top panel). We used a monoclonal antibody that detects an epitope missing in MDM2-C to detect only MDM2-FL (Figure 1C, top panel). We used a commercially available antibody generated against bacterially expressed MDM2 from aa 3–491 to detect both MDM2-FL and MDM2-C (labeled as Pan-MDM2 Figure 1B and C, middle panel). We observed that the monoclonal antibody to the epitope, comprising the 4–10 splice junction, detected only MDM2-C with a mobility of 76 kD (Figure 1B compare top panel lanes 1 and 2). We observed that the monoclonal antibody to the missing 4–10 splice region detected only MDM2-FL at both102 kD and slightly above 76 kD (Figure 1C compare top panel lanes 1 and 2). When the Pan-MDM2 antibody was used that could detect both MDM2-FL and MDM2-C all the forms were observed (middle images of Figure 1B and C). The reason for detecting two bands for MDM2-FL is not known and the Western blot results suggest that the Pan-MDM2 detects the MDM2-FL protein avidly when we compare the stained membrane results to Western blot results (Figure 1A-C). When the detection channels were superimposed, MDM2-C and MDM2-FL showed up as yellow bands, indicating the merging of green and red (bottom images of Figure 1B and C). The antibodies allowed for detection of MDM2-FL and MDM2-C discriminately and verified that we had purified the two different MDM2 isoforms. We previously reported that a form of MDM2-FL migrates very close to MDM2-C on SDS-PAGE gels19 and therefore this verification of the differential antibody detection was very important. We used the Protein Common Interface Database (ProtCID)27 to analyze MDM2-C RING interactions and found an interaction with the E2 ubiquitin-conjugating enzyme UbcH5B and ubiquitin (Figure 1D). This predicted MDM2-C RING interactions with the E2 ubiquitin-conjugating enzyme (UbcH5B) and ubiquitin (Ub) by ProtCID (Protein Common Interface Database).27 The amino acid sequence of MDM2-C was entered and three PFAM IDs were identified: zf-RanBP (Zn-finger in Ran binding protein and others) aa: 129–158; Pinin_SDK_N (pinin/SDK conserved region) aa: 147–255; and zf-C3HC4 (RING finger) aa: 264–315.
MDM2-C Has Auto-Ubiquitination Activity
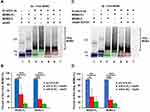
To explore the function of MDM2-C as an E3 ligase, we first asked if MDM2-C had auto-ubiquitination activity. MDM2-FL is known to promote its own ubiquitination.6,25 We predicted that MDM2-C would behave similarly. To test whether MDM2-C could behave as an E3 enzyme for itself we used an in vitro auto-ubiquitination assay and compared MDM2 protein mobility in the presence, or absence, of E1, E2, and ubiquitin (Figure 2). We used a polyclonal antibody to detect both MDM2-C and MDM2-FL (Figure 2A with wtp53 or 2C with mtp53). In the absence of E1, E2, and ubiquitin the MDM2 isoforms migrated as expected (Figure 2A and C, lanes 1 and 2). A major upward shift in size was detected when E1, E2, and ubiquitin were added to the MDM2-C samples. The 76 kD polypeptide was modified and shifted to migrate above 102 kD (Figure 2A and C, lane 3). A similar result was observed for MDM2-FL, which was included as a positive control (Figure 2A and C, lane 4). This demonstrated that MDM2-C had auto-ubiquitination activity. When purified wild-type p53 (Figure 2A wtp53) or mutant p53 (Figure 2C mtp53) was added to the reactions with E1 and E2 an inhibition was observed that resulted in more un-shifted MDM2-C or MDM2-FL band (see Figure 2 quantification by comparing red (+p53) and green bars (-p53) in Figure 2B and D, and compare lanes 5 and 6 to lanes 3 and 4 in 2A and 2C). Competition is expected as different ubiquitination reactions occur at the same time. The histograms constructed to compare non-ubiquitinated MDM2 without E1 and E2 and ubiquitin (Figure 2 blue bars) to non-ubiquitinated MDM2 following a ubiquitination reaction in the presence of wtp53 or mtp53 (Figure 2 red bars) or in their absence (Figure 2 green bars) (Figure 2B and D) we observed that both wtp53 and mtp53 could compete out MDM2-FL and MDM2-C ubiquitination.
MDM2-C Ubiquitinates Wtp53 and Mtp53 R273H
MDM2-FL ubiquitinates wtp53 and mtp53 R273H.4,22,23 The data shown in Figure 2 is the first documentation that MDM2-C has E3 ubiquitination activity. We therefore asked if MDM2-C could ubiquitinate p53 proteins. Previous studies regarding MDM2-C showed that MDM2-C does not lead to robust degradation of wtp53 and MDM2-C expression associates with higher mortality in patients that express mtp53.19,20 Here we tested if the purified MDM2-C could ubiquitinate wtp53 and mtp53. We carried out in vitro p53 ubiquitination assays with the purified wtp53 or mtp53 and Western blot probed for the p53 protein products. As a negative control, we used the bacterial extract and as a positive control, we used MDM2-FL. Upon the addition of either MDM2-FL or MDM2-C, multiple bands above 53 kD appeared in the presence of E1, E2 and ubiquitin (Figure 3A and B, compare lanes 4 and 5 to lanes 1–3). This led us to conclude that MDM2-C not only contained auto-ubiquitination activity but also was able to ubiquitinate wtp53 and mtp53. When we monitored the level of ubiquitination by Image J we observed that MDM2-C significantly shifted both non-ubiquitinated wtp53 and mtp53 to ubiquitinated forms (Figure S2).
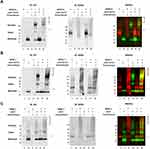
MDM2-C Forms a Complex with Mtp53
We recently reported that MDM2-C expression in breast cancers expressing mtp53 correlates with a worse overall outcome.20 We carried out experiments to determine if MDM2-C, like MDM-FL, was able to form an mtp53 protein-protein complex. The C-terminus of p53 can bind to the N-terminal domain of MDM2.12 Immunoprecipitation experiments showed very slight interactions with a great deal of background so we used the glutaraldehyde cross-linking technique as a way to stabilize any potential interaction (Figure 4). Glutaraldehyde cross-linking can be used to visualize p53 dimers and tetramers.28 We observed that glutaraldehyde cross-linking of MDM2-C did not result in a discrete banding pattern. This was not surprising because it has been observed that the RING domain of MDM2 forms only weak homodimer interactions.29 Therefore, when mixing MDM2-C with either wtp53 or mtp53 we focused our analysis on detecting changes to the p53 protein glutaraldehyde cross-link pattern. The addition of glutaraldehyde shifted the mtp53 and wtp53 pattern from monomers at 53kD to higher migrating oligomers including those of p53 dimers and tetramers (as evidenced by Western blot reactive species above the 102 and 225 kD markers, Figure 4A–C, compare lanes 2 and 4 for IB:anti-p53). When either MDM2-FL or MDM2-C was added to the reaction we detected new mtp53 and wtp53 glutaraldehyde cross-linked protein complexes (as indicated by the lower arrow and upper bracket markers in Figure 4, compare lanes 4 and 6). The bracketed p53 corresponded to cross-linked forms higher than the uppermost p53-alone glutaraldehyde signal in the absence of MDM2 protein (Figure 4A–C anti-p53 lane 4). This was visualized in both the single channel image and as the corresponding merged yellow signal in regions that previously had barely detectable green signal). This strongly suggested the detection of MDM2-C complexed with both wtp53 and mtp53 that was most clearly detectable with mtp53 R273H (Figure 4B, lane 6). The new lower forms of p53 may be the result of a single p53 monomer cross-linked with a single MDM2-C.
Discussion
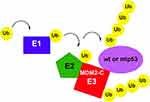
MDM2-C is an alternative spliced form of MDM2 lacking a small portion of the N-terminal p53-binding domain and the nuclear localization signal but it is still able to localize to cancer cell nuclei.19,20 High expression of MDM2-C in breast cancer correlates with a worse overall outcome in cases with co-expression of highly expressed mtp53.18–20 We observed that MDM2-C acts as an E3 ligase for mtp53 and wtp53 (Figures 2, 3 and 5 but at this time do not know how this would influence the p53 proteins in a cellular context. We also found that MDM2-C activated its own auto-ubiquitination (see Figure 5 for model). The degree of ubiquitination by MDM2-C in both auto-ubiquitination and p53 substrate ubiquitination was commensurate to the level of ubiquitination observed by MDM2-FL. MDM2-C was able to interact with both wtp53 and mtp53 as documented by glutaraldehyde cross-linking experiments (Figure 4). It is not yet clear how RING finger E3 ubiquitin ligases target particular lysines to be targets of the ubiquitin thioesters. It remains to be determined which lysines in mtp53, wtp53, and MDM2-C are targets of MDM2-C as well as what is the functional role of MDM2-C mediated ubiquitination.
The interaction between MDM2 and mtp53 sequesters the MDM2 acidic and RING domains and inhibits MDM2 E3 ligase activity.24 We found competitive inhibition of MDM2-C E3 ligase activity by wtp53 or mtp53 as evidenced by reduced MDM2-C auto-ubiquitination (Figure 2). Overall, MDM2-C demonstrated strong auto-ubiquitination E3 ligase activity that was reduced by the addition of either wtp53 or mtp53. These results suggest that MDM2-C acts as an E3 ubiquitin ligase and that its biological activity may be influenced by wtp53 and mtp53 activities. Overexpressed MDM2-B interacts with MDM2-FL and promotes the accumulation of mtp53 by inhibiting p53 degradation.30 The overexpression of MDM2-C in breast cancer shows reciprocal correlations for the mortality outcomes of patients with wtp53 versus mtp53 expression and this may be due to mechanisms associated with increased mtp53 stability. Increased wtp53 causes increased cell death and in spinal muscular atrophy, wtp53 is upregulated by increased Mdm2 exon skipping.31 The overexpression of mtp53 is associated with gained new functions that promote tumorigenesis and metastasis.32 It is highly likely that MDM2-C, like other spliced isoforms of MDM2 promotes mtp53 accumulation and gain-of-function in tumorigenesis.30 The MDM2 splice variant MDM2-B promotes mtp53 protein accumulation in cells by inhibiting MDM2-FL to bind to and degrade mtp53.30 Since MDM2-C has an intact RING domain we predict that MDM2-C can form heterodimers with MDM2-FL through the RING domain that regulates Mdm2-FL function, Future studies are needed to address what proteins MDM2-C may ubiquitinate (in addition to itself and p53) and regulate in vivo. The diverse set of MDM2-C protein–protein interactions, which include interacting with MDM2-FL and mtp53, may participate in context-dependent biological pathways that influence multiple different disease pathways.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi:10.1146/annurev.biochem.78.101807.093809
2. Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–11369. doi:10.1073/pnas.96.20.11364
3. Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi:10.1038/nsmb.2780
4. Fahraeus R, Olivares-Illana V. MDM2’s social network. Oncogene. 2014;33:4365–4376. doi:10.1038/onc.2013.410
5. Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–1008.
6. Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi:10.1074/jbc.275.12.8945
7. Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi:10.1016/S0092-8674(00)81871-1
8. Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi:10.1038/35042675
9. Brooks CL, Gu W. p53 ubiquitination: mdm2 and beyond. Mol Cell. 2006;21:307–315. doi:10.1016/j.molcel.2006.01.020
10. Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem. 2006;281:28575–28583. doi:10.1074/jbc.M513311200
11. Cross B, Chen L, Cheng Q, Li B, Yuan ZM, Chen J. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J Biol Chem. 2011;286:16018–16029. doi:10.1074/jbc.M111.228981
12. Poyurovsky MV, Katz C, Laptenko O, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol. 2010;17:982–989. doi:10.1038/nsmb.1872
13. Nomura K, Klejnot M, Kowalczyk D, et al. Structural analysis of MDM2 RING separates degradation from regulation of p53 transcription activity. Nat Struct Mol Biol. 2017;24:578–587. doi:10.1038/nsmb.3414
14. Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502–9508. doi:10.1158/0008-5472.CAN-05-4271
15. Jacob AG, Singh RK, Comiskey DF
16. Jacob AG, O’Brien D, Singh RK, et al. Stress-induced isoforms of MDM2 and MDM4 correlate with high-grade disease and an altered splicing network in pediatric rhabdomyosarcoma. Neoplasia. 2013;15:1049–1063. doi:10.1593/neo.13286
17. Rosso M, Okoro DE, Bargonetti J. Splice variants of MDM2 in oncogenesis. Subcell Biochem. 2014;85:247–261.
18. Okoro DR, Rosso M, Bargonetti J. Splicing up mdm2 for cancer proteome diversity. Genes Cancer. 2012;3:311–319. doi:10.1177/1947601912455323
19. Okoro DR, Arva N, Gao C, et al. Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS One. 2013;8:e77643. doi:10.1371/journal.pone.0077643
20. Loo LWM, Gao C, Shvetsov YB, Okoro DR, Hernandez BY, Bargonetti J. MDM2, MDM2-C, and mutant p53 expression influence breast cancer survival in a multiethnic population. Breast Cancer Res Treat. 2018.
21. Fan C, Wang X. Mdm2 Splice isoforms regulate the p53/Mdm2/Mdm4 regulatory circuit via RING domain-mediated ubiquitination of p53 and Mdm4. Cell Cycle. 2017;16:660–664. doi:10.1080/15384101.2017.1288327
22. Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi:10.1128/MCB.00050-07
23. Li D, Marchenko ND, Schulz R, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi:10.1158/1541-7786.MCR-10-0534
24. Yang L, Song T, Cheng Q, Chen L, Chen J. Mutant p53 sequestration of the MDM2 acidic domain inhibits E3 ligase activity. Mol Cell Biol. 2019;39.
25. Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi:10.1038/sj.emboj.7600145
26. Bargonetti J, Friedman PN, Kern SE, Vogelstein B, Prives C. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991;65:1083–1091. doi:10.1016/0092-8674(91)90560-L
27. Xu Q, Dunbrack RL
28. Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci U S A. 1993;90:3319–3323. doi:10.1073/pnas.90.8.3319
29. Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi:10.1016/S0014-5793(99)00254-9
30. Zheng T, Wang J, Zhao Y, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi:10.1038/ncomms3996
31. Van Alstyne M, Simon CM, Sardi SP, Shihabuddin LS, Mentis GZ, Pellizzoni L. Dysregulation of Mdm2 and Mdm4 alternative splicing underlies motor neuron death in spinal muscular atrophy. Genes Dev. 2018;32:1045–1059. doi:10.1101/gad.316059.118
32. Bargonetti J, Prives C. Gain-of-function mutant p53: history and speculation. J Mol Cell Biol. 2019;11:605–609. doi:10.1093/jmcb/mjz067
33. Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.