Back to Journals » OncoTargets and Therapy » Volume 13
Matrix Metalloproteinase 11 is a Potential Biomarker in Bladder Cancer Diagnosis and Prognosis
Authors Chen C, Liu X, Jiang J, Li S, Wang G, Ju L , Wang F , Liu T, Li S
Received 23 December 2019
Accepted for publication 16 August 2020
Published 11 September 2020 Volume 2020:13 Pages 9059—9069
DOI https://doi.org/10.2147/OTT.S243452
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Leo Jen-Liang Su
Chen Chen,1,2 Xiaoping Liu,3 Jiazhi Jiang,3 Shenjuan Li,1,2 Gang Wang,1,2 Lingao Ju,1,2 Fubing Wang,1,2 Tongzu Liu,3 Sheng Li1– 3
1Department of Biological Repositories, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China; 2Human Genetics Resource Preservation Center of Wuhan University, Wuhan, People’s Republic of China; 3Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China
Correspondence: Sheng Li
Department of Biological Repositories, Zhongnan Hospital of Wuhan University, 169 Donghu Road, Wuhan 430071, People’s Republic of China
Email [email protected]
Purpose: Bladder cancer is one of the leading causes of cancer death all over the world, and half of patients are diagnosed at advanced stages with poor therapeutic response. Thus, developing new biomarkers for bladder cancer diagnosis and prognosis is urgently needed.
Materials and Methods: Bioinformatic and gene ontology (GO) analysis were employed to screen highly upregulated and secretory tumor markers in the TCGA BLCA cohort. IHC in tissue microarray and ELISA in cancer cell culture medium were used to validate the expression of putative biomarkers in bladder cancer. Bisulfite sequencing was used to detect DNA methylation status in the promoter of putative genes.
Results: In this study, MMP11 is first identified as one of the most differentially expressed genes (DEGs) in bladder cancer by meta-analysis in a TCGA bladder cancer cohort. The strong upregulation of MMP11 is confirmed at protein levels in both bladder cancer patients and cell lines. Mechanistic studies reveal that MMP11 promoter hypomethylation, but not genomic amplification or mutation, accounts for its enhanced expression in bladder cancer both in vitro and in vivo. Moreover, clinicopathological analysis indicates that MMP11 upregulation is associated with the tumor progression and poor survival in bladder cancer patients.
Discussion: These findings suggest that MMP11, as a secretory protein, is a promising biomarker for diagnosis and prognosis in bladder cancer.
Keywords: bladder cancer, TCGA, MMP11, DEG, hypomethylation, diagnosis, prognosis
Introduction
Bladder cancer is the most common cancer of the urinary tract, with an estimated 80,470 new cases and 17,670 deaths projected to occur in the United States in 2019.1 Due to the advances in early detection and treatment for bladder cancer, the 5-year overall survival rate increases to 90%, whereas 40–80% of these patients will eventually develop recurrence of disease or progression.2,3 At present, cystoscopy and urine cytology are the most reliable methods for bladder cancer diagnosis.4 However, the invasive diagnostic procedure in cystoscopy often leads to discomfort and distress in patients. Urine cytology has high specificity but low sensitivity for low-grade bladder cancer.5 Although several other methods were approved by FDA, including BTAstat, BTAtrak, NMP22, FDP, ImmunoCyt, and FISH,6 none of them can replace cystoscopy and urine cytology in bladder cancer detection due to their low sensitivity or poor specificity. Thus, to develop a highly sensitive, specific, non-invasive, painless, low-cost, and long-term follow-up method has become a hot spot in bladder cancer.
Previous studies by our group and other researchers have identified several deregulated genes and signaling pathways in BCa and normal bladder tissues.7,8 In a small RNA-seq cohort of bladder cancer, 17 hub genes including MMP11 were found to be associated with TNM staging and overall survival, which is correlated with the data from Li et al9 showing the prognostic role of MMP11 in metastasis and poor survival in urothelial carcinomas by IHC study.
Matrix metalloproteinases (MMPs) are extracellular enzymes which break down the ECM and regulate cytokine and growth factor activity.10–13 MMP11, also known as stromelysm-3, a member of the stromelysin subgroup belonging to the MMPs superfamily, was initially identified in invasive breast carcinoma.14 In physiological condition, MMP11 is involved in tissue remodeling during embryogenesis,15 tissue involution, wound healing,16 and metamorphosis.17 Meanwhile, MMP-11 is highly expressed in various cancers, including oral cancer,18,19 desmoid tumors,20 lung cancer,21 esophageal carcinoma,22 pancreatic carcinoma,23 aggressive meningioma,24 ovarian carcinoma,25 and colon cancer.26 Although some progress has been made in MMP11’s prognosis role in bladder cancer, its diagnosis role and underlying mechanism are unknown.27
Here, our systemic study shows that MMP11 is highly expressed both in Bladder cancer tissue and cell lines, which is because of its promoter hypomethylation. Clinicopathological analysis indicates MMP11 is correlated with poor clinical outcomes and survival of patients with bladder cancer. Given its secretory property, MMP11 is a promising biomarker for bladder cancer diagnosis and prognosis.
Materials and Methods
Data Collection and Data Processing
Bladder cancer gene expression RNAseq (IlluminaHiSeq pancan normalized) were downloaded from the TCGA BLCA cohort (http://xena.ucsc.edu/). After excluding unqualified samples, a total of 407 bladder cancer samples and 19 normal samples were used for the following analysis. The normalized and log2-transformed RNA-seq data displayed as count number were conducted by using R package “DEseq.2” to screen differentially expressed genes (DEGs) in bladder cancer.28 The data about gene methylation, mutation, copy number variation, and clinicopathological features (including sex, pathologic stage, T stage, N stage, metastasis, recurrence, histological grade, diagnosis subtype, and survival) were also retrieved from the TCGA BLCA cohort. In the present study, R package “maxstat” was employed for grouping samples to generate maximal difference in patient overall survival. Meanwhile, the gene expression profile that MMP11 is strongly upregulated in cancer tissues was also considered as a critical factor to group samples. Thus, 0.28 was set as the optimal cutoff to meet both criteria mentioned above for grouping samples (Figure S1, Table 1). In detail, bladder cancer patients in the TCGA BLCA cohort were divided into a high-expression group (n=262) and low-expression group (n=145) (Supplementary Figure S1, Table 1). Association between MMP11 and clinicopathological parameters in bladder cancer was examined by Chi-square (χ2) test or Fisher’s exact test in GraphPad Prism 8. Overall survival and recurrence free survival were analyzed by Kaplan-Meier curve in GraphPad Prism 8. Log rank test was used to determine the significance of difference in survival analysis. Student’s t-test (two tailed, unpaired unless otherwise specified) was used to assess significance of differences between two groups. All bars in figures represent means±SEM. The P-values are indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ns, not statistically significant.
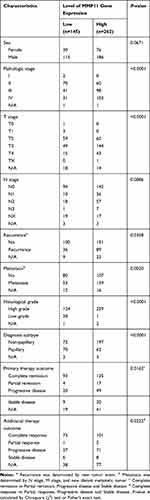 |
Table 1 Clinicopathological Characteristics in TCGA BLCA Cohort |
Univariate and Multivariate Cox Regression Analysis
Univariate and multivariate Cox regression analysis were performed to determine the association of MMP11 with patient overall survival by SPSS (version 21.0). All date including MMP11 expression values and clinical features (age, sex, pathologic stage, T stage, N stage, M stage, and histological grade) were downloaded from a TCGA BLCA cohort. Clinical feathers with P<0.05 were used for multivariate Cox analysis to adjust the correlation between MMP11 and overall survival.
Differentially Expressed Genes (DEGs) Screening and Gene Ontology (GO) Term Analysis
Gene expression level was compared between bladder cancer tissues (n=407) and adjacent normal tissues (n=19) to screen differentially expressed genes (DEGs) in TCGA-BLCA by R package “edgeR”28 P-value<0.05 and |log2FC|≥1 were set as cutoff values, which was in accordance to that in most bioinformatic analysis in the literature.29,30 DEGs had further gene ontology (GO) term cellular component analysis conducted at http://www.geneontology.org/ to identify secretory biomarkers.
Cell Culture
Bladder cancer cell lines (T24, UMUC3, and 5637) and normal bladder cell line SV-HUC-1 were maintained at 5% CO2, 37°C in RPMI 1640 medium supplemented with 10% FBS. Commercialized cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
Enzyme-Linked Immunosorbent Assay (ELISA)
Culture medium from 106 cells seeded for 48 hours was collected in a sterile container and then centrifuged at a speed of 2000/3000 rpm for 20 minutes. The supernatant was recovered and subjected to protein detection by MMP11 ELISA kits (m1058665; Mlbio, China). The optimal detection range was 5~200 ng/mL. To generate a standard curve, serial dilutions of standard material provided in ELISA kit were performed to obtain protein solutions at the concentrations of 150 ng/mL, 100 ng/mL, 50 ng/mL, 25 ng/mL, and 12.5 ng/mL. Meanwhile, experimental samples from supernatant of cultured cells were seeded in the same plate. Blank wells without samples or ELISA reagents were also included. Then absorbance (OD value) at 450 nm wavelength was measured in each well within 15 minutes. All samples in one experiment were analyzed in triplicate and the experiment was repeated at least three times.
DNA Methylation
Genomic DNA from bladder cancer cell lines (T24, UMUC3, and 5637) and normal bladder cell line SV-HUC-1 was extracted by QIAamp DNA Mini Kit), and treated with sodium bisulfite according to the manufacturer’s protocol (51034, Qiagen, NO:59104, Germany). Then, the resulting DNA was PCR amplified with forward primer (TGGGGGAATTTGTTTGTGGGTAG) and reverse primer (CCCCTTATAGCTTCCTCCCCAC) by 2×Es Taq MasterMix in the reaction: 95°C for 5 minutes, and 45 cycles of 95°C for 30 seconds, 52°C for 30 seconds, 72°C for 30 seconds. The purified PCR products were cloned into plasmid and sequenced to determine the methylation status at the following CpG sites of MMP11 gene in MMP11 promoter: −347CpG (cg22418565), −267CpG (cg23850277), −180CpG (cg14420645), −129CpG (cg21246624), and −66CpG (cg13361393). PCR reactions were performed using 2×Es Taq MasterMix at 95°C for 5 minutes, and 45 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 30 seconds. For each cell line, 30 clones were randomly selected for sequencing for each cell line in one experiment, and the data shown in Figure 1E were the pool of three independent experiments.
Immunohistochemistry in Tissue Microarray
A bladder cancer tissue microarray with 30 cancer tissues and 30 paracancerous tissues was purchased from the Shanghai Outdo Biotech Company (Shanghai China), and subjected to MMP11 protein detection by IHC. Briefly, TMA sections underwent deparaffinization, rehydration antigen retrieval in citrate buffer (pH 6.0) at >90°C for 15 minutes, and endogenous peroxidase quench by 3% H2O2 at room temperature for 15 minutes. After blocking, the sections were incubated with primary antibody (1:400, CSB-PA003254, CusAB), and visualized by EnVision FLEX+ Systems (K8002, DAKO) under microscope. MMP11 protein level was quantified as histochemical score (H-score) by two pathologists. Briefly, the percentage of positive cell in each sample was scored as follows: 0 points for <5%, 1 point for 5%~25%, 2 points for 26%~50%, 3 points for 51%~75%, 4 points for >75%. The intensity of positive staining was scored as follows: 0 points for colorless, 1 point for light yellow, 2 points for brown yellow, and 3 points for dark brown. MMP11 protein level was obtained by multiplying percentage score and intensity score, and further defined as follows: 0 for negative, 1~4 for weak, 5~8 for moderate, and 9~12 for strong. Scores were averaged in both cancer tissue and border tissues (Figure 2B), and the clinicopathological characteristics are summarized in Supplementary Table S2. The data in patient tissues are from the public resource TCGA BLCA cohort, commercialized bladder cancer tissue microarray and cell lines, purchased from a Chinese Company, so they do not raise ethical issues. The Ethics Committee at Zhongnan Hospital of Wuhan University approved the study (approval number: 2015029).
Results
Big Data Analysis Reveals MMP11 Overexpression in Bladder Cancer
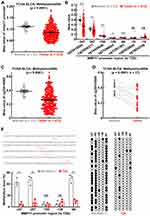
To identify a putative biomarker for bladder cancer diagnosis, big data analysis was conducted in gene expression profile in TCGA BLCA cohort and 4442 DEGs (P-value<0.05 and |log2FC|≥1) were retrieved from a total of 20,530 genes (Figure 3A). GO term cellular component analysis was performed for these DEGs (1712 upregulated and 2730 down-regulated), and revealed that MMP11 is one of the most differentially expressed secretory proteins in bladder cancer (Figure 3A and B).To verify this finding, the data about MMP11 were retrieved from a TCGA BLCA cohort. As expected, a ~14.9-fold increase of MMP11 expression is observed in bladder tumor tissues compared to adjacent normal tissues (Figure 3C), whereas a higher upregulation of MMP11 expression (28.7-fold) is found in bladder cancer tissues compared to their matched normal tissues (Figure 3D). In addition, MMP11 expression level gradually increased along with patient tumor stage, which suggests its role in cancer progression (Figure 3E). Together, these data suggest that MMP11 is highly overexpressed in bladder cancer.
MMP11 is Upregulated in Bladder Cancer
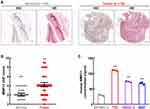
To verify the upregulation of MMP11 in bladder cancer, IHC was first employed to detect MMP11 protein level in a bladder cancer tissue microarray. As expected, the MMP11 protein level is significantly higher in bladder cancer tissues compared to their adjacent normal tissues (Figure 2A and B, Supplementary Table S2). Since MMP11 is a secretory protein, ELISA was also introduced to detect the MMP11 protein level in culture medium from bladder cancer cell lines. As shown in Figure 2C, compared with normal bladder cell SV-HUC-1, bladder tumor cells (T24, UMUC3, and 5637) secreted much more MMP11 protein into extracellular space. Taken together, these data strongly indicate that MMP11 is upregulated in bladder cancer, conferring it is a potential biomarker for bladder cancer diagnosis.
Promoter Hypomethylation Contributes to MMP11 Upregulation in Bladder Cancer
To determine the underlying mechanism of MMP11 upregulation in bladder cancer, the genomic information of the MMP11 gene was retrieved from the TCGA BLCA cohort. Neither mutation nor amplification was found in the MMP11 gene locus (Supplementary Figure S3). However, compared to normal bladder tissues, a significant decrease in CpG methylation was found in the MMP11 promoter region in bladder cancer tissues, in particular at −347CpG site (Figure 1A–D). Consistently, bladder cancer cell T24 also exhibited remarkably lower methylation levels at −347CpG, −267CpG, and −66CpG sites in MMP11 promoter than that in normal bladder cell SV-HUC-1 (Figure 1E). These data together strongly suggest that promoter hypomethylation accounts for MMP11 upregulation in bladder cancer.
MMP11 Overexpression is Associated with Tumor Progression and Poor Patient Survival in Bladder Cancer
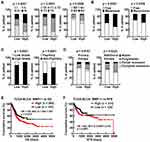
Cancer cell utilizes promoter methylation as one essential epigenetic tool to silence tumor suppressor gene or activate oncogene to promote tumorigenesis. Given increased upregulation of MMP11 along with tumor stage (Figure 3E) and hypomethylation in MMP11 promoter (Figure 1), MMP11 is likely to play a pro-tumorigenic role in bladder cancer progression. To prove this hypothesis, we evaluated the correlation between MMP11 expression and clinicopathological factors in bladder cancer patients from the TCGA BLCA cohort. As shown in Table 1, there is no significant difference between gender and MMP11 expression level in bladder cancer patients. However, a high MMP11 expression level is positively associated with advanced tumor stage (pathological stage, T stage and N stage, P<0.01), recurrence (P=0.0358), metastasis (P=0.0020), high disease grade (P<0.0001), and non-papillary subtype (P<0.0001) (Figure 4A–C, Table 1). Moreover, a negative correlation was found between MMP11 expression level and therapeutic efficacy in bladder cancer (primary therapy outcome: P=0.0163; additional therapeutic outcome: P=0.0222) (Figure 4D, Table 1). Consistent with its tumor-promoting role in bladder cancer progression and therapeutic response, MMP11 overexpression is associated with poor overall survival (HR=1.419, 95% CI=1.040–1.937, P=0.0275) and poor recurrence free survival (HR=1.532, 95% CI= 0.9897–2.720, P=0.0557) in bladder cancer patients (Figure 4E and F). Taken together, these data indicate that MMP11 upregulation is associated with tumor progression and patient survival, conferring its a potential biomarker in bladder cancer prognosis.
Discussion
Bladder cancer is a malignant disease with high incidence and mortality rate. About half of bladder cancer patients were diagnosed at advanced stage with poor therapeutic response and decreased survival rate, while the remaining cases in early stage (non-invasive) had a significantly increased survival rate. Thus, to develop a novel diagnostic biomarker is urgently needed for bladder cancer patients.31
To screen the putative tumor marker for bladder cancer diagnosis, big data mining and bioinformatic analysis were performed in the TCGA BLCA cohort. In total, 4442 DEGs were identified (1712 upregulated and 2730 downregulated). Among these genes, MMP11 is one of the most upregulated genes in bladder cancer patients, and specifically in all BLCA subtypes except luminal papillary: (neuronal, n=20, P<0.0001; basal squamous, n=142, P<0.0001; luminal, n=26, P<0.0001, luminal infiltrated, n=78, P< 0.0001; luminal papillary, n = 142, P=0.1006) (Figure 3, Supplementary Figure S2), which is confirmed by previous studies showing the prognostic role of MMP11 and another 10 genes in bladder cancer progression and survival.9,10 Given that current diagnostic procedures, such as tissue examination and urine cytology, are invasive or insensitive for bladder cancer detection, particularly for those in early stages, MMP11 was selected as a putative diagnostic biomarker for further study due to its secretory property and high upregulation (Figure 3). Indeed, a strongly elevated MMP11 protein level was detected in both primary bladder cancer tissues and culture medium from bladder cancer cell lines (Figure 2). These results render MMP11 promising in bladder cancer diagnosis.
Tumor cells utilize multiple mechanisms to regulate the expression of oncogene and tumor suppressor gene to promote tumor initiation, progression, and therapeutic resistance, for example, mutation, amplification/deletion, DNA methylation, and histone deacetylation. In the present study, promoter hypomethylation, but not mutation or amplification, is involved in MMP11 upregulation in bladder cancer both in vivo and in vitro (Figure 1, Supplementary Figure S3). These data suggest that the methylation status in MMP11 promoter can also be used as a diagnostic and prognostic indicator in bladder cancer. Although much more time, effort, and patient tissues are required for this complicated procedure, the information about DNA methylation in MMP11 promoter can provide additional evidences for bladder cancer diagnosis and prognosis.
MMP11, like other members in the MMP family, functions as a matrix metallopeptidase to regulate the extracellular matrix (ECM) protein levels, suggesting a critical role in solid malignant tumor migration and invasion.26,32 In 2016, Li et al10 defined a prognostic role of MMP11 in metastasis and poor survival in urothelial carcinomas by IHC study. Later on, our previous study in bladder cancer RNA-seq identified 11 genes including MMP11 associated with TNM staging and patient survival. Similarly, MMP11 was confirmed to be positively associated with tumor stage (pathological, T, and N), metastasis and recurrence, and tumor grade, but negatively with therapeutic outcome and survival in the TCGA BLCA cohort (Figure 4, Table 1). In addition, the correlation between MMP11 and patient overall survival was validated by Cox regression analysis (HR=1.057, 95% CI=0.995~1.123, P=0.028), although only a trend of correlation was detected when adjusted by the indicated clinical features (Supplementary Table S1). Hence, besides the diagnostic role, MMP11 can also function as a prognostic biomarker in bladder cancer.
The prognostic role of MMP11 in bladder cancer is probably mediated by its pro-tumorigenic activity. Thus, the future direction will focus on the function of MMP11 in bladder cancer progression and therapeutic resistance. Given that MMP11 protein is predominantly located in the extracellular matrix, the inhibition targeting will be feasible by specific inhibitor or neutralizing antibody, which eventually changes the tumor microenvironment to benefit cancer therapies, including chemotherapy and immunotherapy.
The comprehensive study demonstrated that promoter hypomethylation-caused strong upregulation of MMP11 in bladder cancer is associated with worse clinical outcomes. With more data on patients, ELISA detection of MMP11 protein level in urine will be a fast, sensitive, and noninvasive method for bladder cancer diagnosis and prognosis.
Data Sharing Statement
The gene profile about expression, methylation, mutation, copy number, and related clinical data of bladder cancer were retrieved from The Cancer Genome Atlas (TCGA) database (http://xena.ucsc.edu/).
Acknowledgments
This work was supported by the Fundamental Research Fund for the Central Universities (2042019kf0160), the Zhongnan Hospital of Wuhan University Science, Technology and Innovation Cultivating Fund (cxpy2019077) and the Wuhan Youth Talent Project (WHQG201901).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Ca Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Malmstrom PU, Agrawal S, Blackberg M, et al. Non-muscle-invasive bladder cancer: a vision for the future. Scand J Urol. 2017;51(2):87–94. doi:10.1080/21681805.2017.1283359
3. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. Ca Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
4. Shariat SF, Karam JA, Lotan Y, Karakiewizc PI. Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev Urol. 2008;10(2):120–135.
5. Kiyoshima K, Akitake M, Shiota M, et al. Prognostic Significance of Preoperative Urine Cytology in Low-grade Non-muscle-invasive Bladder Cancer. Anticancer Res. 2016;36(2):799–802.
6. Mohammed AA, El-Tanni H, El-Khatib HM, Mirza AA, Alturaifi TH. Urinary Bladder Cancer: biomarkers and Target Therapy, New Era for More Attention. Oncol Rev. 2016;10(2):10.
7. Cao R, Wang G, Qian K, et al. TM4SF1 regulates apoptosis, cell cycle and ROS metabolism via the PPARgamma-SIRT1 feedback loop in human bladder cancer cells. Cancer Lett. 2018;414:278–293. doi:10.1016/j.canlet.2017.11.015
8. Wang G, Cao R, Wang Y, et al. Simvastatin induces cell cycle arrest and inhibits proliferation of bladder cancer cells via PPARgamma signalling pathway. Sci Rep. 2016;6(1):35783. doi:10.1038/srep35783
9. Li S, Liu X, Liu T, et al. Identification of Biomarkers Correlated with the TNM Staging and Overall Survival of Patients with Bladder Cancer. Front Physiol. 2017;8:947. doi:10.3389/fphys.2017.00947
10. Li WM, Wei YC, Huang CN, et al. Matrix metalloproteinase-11 as a marker of metastasis and predictor of poor survival in urothelial carcinomas. J Surg Oncol. 2016;113(6):700–707. doi:10.1002/jso.24195
11. Boulay A, Masson R, Chenard MP, et al. High cancer cell death in syngeneic tumors developed in host mice deficient for the stromelysin-3 matrix metalloproteinase. Cancer Res. 2001;61(5):2189–2193.
12. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. doi:10.1038/nrc745
13. Matziari M, Dive V, Yiotakis A. Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med Res Rev. 2007;27(4):528–552. doi:10.1002/med.20066
14. Basset P, Bellocq JP, Wolf C, et al. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 1990;348(6303):699–704. doi:10.1038/348699a0
15. Lefebvre O, Regnier C, Chenard MP, et al. Developmental expression of mouse stromelysin-3 mRNA. Development. 1995;121(4):947–955.
16. Okada A, Saez S, Misumi Y, Basset P. Rat stromelysin 3: cDNA cloning from healing skin wound, activation by furin and expression in rat tissues. Gene. 1997;185(2):187–193. doi:10.1016/S0378-1119(96)00615-4
17. Wang Z, Brown DD. Thyroid hormone-induced gene expression program for amphibian tail resorption. J Biol Chem. 1993;268(22):16270–16278.
18. Arora S, Kaur J, Sharma C, et al. Stromelysin 3, Ets-1, and vascular endothelial growth factor expression in oral precancerous and cancerous lesions: correlation with microvessel density, progression, and prognosis. Clin Cancer Res. 2005;11(6):2272–2284. doi:10.1158/1078-0432.CCR-04-0572
19. Soni S, Mathur M, Shukla NK, Deo SV, Ralhan R. Stromelysin-3 expression is an early event in human oral tumorigenesis. Int J Cancer. 2003;107(2):309–316. doi:10.1002/ijc.11366
20. Denys H, De Wever O, Nusgens B, et al. Invasion and MMP expression profile in desmoid tumours. Br J Cancer. 2004;90(7):1443–1449. doi:10.1038/sj.bjc.6601661
21. Kettunen E, Anttila S, Seppanen JK, et al. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149(2):98–106. doi:10.1016/S0165-4608(03)00300-5
22. Hourihan RN, O’Sullivan GC, Morgan JG. Transcriptional gene expression profiles of oesophageal adenocarcinoma and normal oesophageal tissues. Anticancer Res. 2003;23(1A):161–165.
23. von Marschall Z, Riecken EO, Rosewicz S. Stromelysin 3 is overexpressed in human pancreatic carcinoma and regulated by retinoic acid in pancreatic carcinoma cell lines. Gut. 1998;43(5):692–698. doi:10.1136/gut.43.5.692
24. Perret AG, Duthel R, Fotso MJ, Brunon J, Mosnier JF. Stromelysin-3 is expressed by aggressive meningiomas. Cancer. 2002;94(3):765–772. doi:10.1002/cncr.10270
25. Mueller J, Brebeck B, Schmalfeldt B, Kuhn W, Graeff H, Hofler H. Stromelysin-3 expression in invasive ovarian carcinomas and tumours of low malignant potential. Virchows Arch. 2000;437(6):618–624. doi:10.1007/s004280000261
26. Wlodarczyk J, Stolte M, Mueller J. E-cadherin, beta-catenin and stromelysin-3 expression in de novo carcinoma of the colorectum. Pol J Pathol. 2001;52(3):119–124.
27. Zhang X, Huang S, Guo J, et al. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review). Int J Oncol. 2016;48(5):1783–1793. doi:10.3892/ijo.2016.3400
28. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi:10.1093/bioinformatics/btp616
29. Li MX, Jin LT, Wang TJ, et al. Identification of potential core genes in triple negative breast cancer using bioinformatics analysis. Onco Targets Ther. 2018;11:4105–4112. doi:10.2147/OTT.S166567
30. Sun C, Yuan Q, Wu D, Meng X, Wang B. Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget. 2017;8(41):70271–70280. doi:10.18632/oncotarget.20082
31. Clark PE, Spiess PE, Agarwal N, et al. NCCN Guidelines Insights: bladder Cancer, Version 2.2016. J Natl Compr Canc Net. 2016;14(10):1213–1224. doi:10.6004/jnccn.2016.0131
32. Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124(4):619–626. doi:10.1083/jcb.124.4.619
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.