Back to Journals » Nature and Science of Sleep » Volume 14
Manuscript Title: A 4-miRNAs Serum Panel for Obstructive Sleep Apnea Syndrome Screening
Authors Mo J, Zeng C, Li W, Song W, Xu P
Received 18 July 2022
Accepted for publication 1 November 2022
Published 9 November 2022 Volume 2022:14 Pages 2055—2064
DOI https://doi.org/10.2147/NSS.S382765
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Ahmed BaHammam
Jianming Mo,* Chao Zeng,* Wei Li,* Weidong Song, Ping Xu
Department of Pulmonary and Critical Care Medicine, Peking University Shenzhen Hospital, Shenzhen, 518034, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ping Xu, Peking University Shenzhen Hospital, 1120 Lianhua Road, Shenzhen, 518036, People’s Republic of China, Email [email protected]
Background: Obstructive sleep apnea syndrome (OSAS) is a common chronic sleep disorder. OSAS is closely related to cardiovascular disease, metabolic disorders, cancer risk, and sudden death. This association has special significance in young people. Although it is known that OSAS has a great impact on physical health, it is estimated that 70– 80% of patients with moderate-to-severe OSAS remain undiagnosed. Therefore, a new method for early diagnosis of the disease, the therapeutic effect of OSAS and prevention of complications to important.
Methods: A total of 110 patients with moderate-to-severe OSAS diagnosed in the Sleep Disorders Diagnosis and Treatment Center of Peking University Shenzhen Hospital were selected. After excluding other diseases, 59 patients were finally selected as the OSAS group. In addition, 60 healthy people were selected as the control group. Serum RNA was then extracted. Eight RNA samples were randomly selected from the two groups for high-throughput miRNA sequencing. The 10 miRNAs with the greatest differences were selected as preselected markers from the results. Then, qRT-PCR was performed on the remaining RNA samples of the two groups to extract and verify the 10 miRNAs, and statistical analysis was performed between groups.
Results: A diagnostic panel was constructed by a stepwise logistic regression model combined with the expression data of miRNAs in the validation phase. A four-miRNA panel was identified to better predict OSAS, and the model was calculated using the following formula: Logit (P)= 0.77– 1.65 × miR-486-5p - 4.56 × miR-148a-3p + 1.79 × miR-744-5p + 1.13 × let-7d-3p. The AUC for the four-miRNA panel was 0.955 (95% CI: 0.899 to 0.985; sensitivity = 91.38%, specificity = 91.38%). Gene Ontology (GO) annotation and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis was included in bioinformatic analysis.
Conclusion: A 4-miRNAs panel as a diagnostic biomarker for OSAS screening is feasible.
Keywords: biomarkers, microRNA, obstructive sleep apnea syndrome, diagnosis, bioinformatics
Introduction
Obstructive sleep apnea syndrome (OSAS) is a common chronic sleep disorder.1 Symptoms such as oxygen desaturation and daytime sleepiness occur in approximately 10–17% of the adult population. OSAS is characterized by recurrent episodes of obstruction of the upper airway during sleep, causing recurrent episodes of chronic intermittent hypoxia (CIH), which lead to sleep fragmentation and daytime sleepiness. In obstructive airway conditions, the ineffective effects of breathing exercise also led to increased chest pressure, intermittent suffocation, and disruption of the sleep-wake cycle. Taken together, these events strongly associate OSAS with the risk of cardiovascular disease, metabolic disorders, cancer development, and sudden death, an association of particular significance in the younger population.2 In recent years, the incidence of cancer in young adults has increased, in which OSAS may be involved and play a larger role. In addition, CIH may also cause oxidative stress, inflammation, etc., and even the delay in OSAS treatment will have irreversible effects on cardiovascular disease. Although it is known that OSAS has a great impact on physical health, it is estimated that 70–80% of patients with moderate-to-severe OSAS remain undiagnosed.3 Therefore, a new method for early diagnosis of the disease, the therapeutic effect of OSAS and the prevention of complications to important.
Traditional polysomnography (PSG) is the most important diagnostic method for OSAS. Through continuous monitoring of breathing, arterial oxygen saturation, electroencephalogram (EEG), electrocardiogram (ECG), heart rate and electromyogram (EMG) during sleep, doctors can make a clear diagnosis based on the analysis of the monitoring data. But PSG requires seven hours of continuous testing while the patient is asleep, and the wires that are connected can easily come off. Therefore, data loss and errors are often caused. In addition, in many patients, the lead severely affects the quality of sleep, leading to an inaccurate diagnosis of OSAHS.4 These directly lead to the difficulty of OSAS screening. Often when the disease is diagnosed, the degree of nocturnal apnea and hypoxia of the patient is already very serious, and irreversible cardiovascular disease has occurred. Therefore, high-throughput sequencing or qRT-PCR diagnostic methods can improve the underdiagnosis of diseases and can reduce unnecessary procedures, thereby reducing the use of medical resources.
Currently, research in the field of OSAS is focused on finding its reliable biomarkers, which can not only help doctors make a quick diagnosis, but also increase the understanding of the physiopathological genetic level of OSAS.5 In the era of precision medicine, miRNAs have become an increasingly common research object for the screening, diagnosis and management of various diseases. miRNAs are a class of small noncoding RNAs that negatively regulate gene expression post-transcriptionally by binding to target messenger RNAs (mRNAs), resulting in degradation or translational repression and protein synthesis.6 They play key roles in several biological processes, such as stress response, apoptosis, proliferation and differentiation, and many studies have shown that they are overexpressed in several diseases including cardiovascular disease, metabolic disorders and cancer. miRNAs are ubiquitous in blood, are resistant to degradation, and can be measured simply and quickly (8–10 hours), making them ideal biomarkers.7
This study aimed to detect miRNA expression in serum, identify differences between OSAS patients and healthy people, and explore their clinical significance and contribution to disease diagnosis. In addition, we combined the NoSAS score to try to find a simpler and more reliable clinical diagnosis method.
Materials and Methods
Participants Enrollment and Serum Collection
From January 2020 to February 2021, a total of 110 patients with moderate-to-severe OSAS diagnosed at the Sleep Disorders Diagnosis and Treatment Center of Peking University Shenzhen Hospital were selected. All patients underwent Philips polysomnography (Alice6LDE 31-lead PSG). 1 night (≥ 7 hours) of continuous sleep monitoring, and the monitoring results met the OSAS diagnostic criteria of the American Academy of Sleep Medicine (AASM).8 Exclusion criteria: (1) Combined with other sleep disorders; (2) Combined with chronic or severe cardiopulmonary diseases; (3) Combined with cerebrovascular disease, diabetes, hyperthyroidism, dementia, anxiety and depression or mental disorders; (4) Combined with tumors (5) Combined pregnancy; (6) Received continuous positive airway pressure (CPAP) or surgical treatment; (7) Taking psychotropic and sedative drugs; (8) History of smoking and alcoholism. Finally, 59 patients were selected as the OSAS group. During the same period, 60 healthy people were recruited and selected as the control group. OSAS were excluded by the same polysomnography, and they met the above exclusion criteria. Two groups of patients took fasting whole blood samples from 8:00–9:00 in the morning and centrifuged them to obtain serum. The serum was collected and stored at −80°C for subsequent batch extraction of RNA. RNA was extracted by TRIZOL method (Trizol, THERMO 15596026). The basic data of the two groups of patients are shown in Table 1.
 |
Table 1 The Clinical Characteristics of Participants |
Candidate miRNA Screening
Build the library: Eight RNA samples were randomly selected from the two groups, the RNA concentration was accurately quantified by Qubit, and the integrity of RNA was accurately detected by Agilent 2100. Libraries were constructed using the Small RNA Sample Pre Kit.
miRNA screening: After the sample library is qualified, high-throughput miRNA sequencing (illumina HiSeqTM2500/MiSeq sequencing platform) is performed, and the raw reads of the original sequencing results are filtered to obtain “Clean Reads”, and then the length distribution statistics of “Clean Reads” are performed and screened out SmallRNA (sRNA) within the length range of miRNA (21~22nt).
The sRNA after length screening is located on the reference sequence, the distribution of sRNA on the reference sequence is analyzed, and the density statistics of all the “Reads” of each sample aligned to each chromosome on the genome are performed, and the circos plot is used to view each chromosome. The distribution of Reads is “Mapped Reads”. Align the “Reads” on the above-mentioned “Mapped Reads” reference sequence with the specified range sequence in miRbase to obtain the details of the matched sRNAs in each sample, including the secondary structure of the matched known miRNAs, the sequence, length, number of occurrences and other information of miRNA in each sample.
Differential analysis of miRNA: The expression levels of known and new miRNAs in each sample were counted, and the expression levels were normalized with TPM. The input data of miRNA differential expression is the “Readcount” data obtained in the analysis of miRNA expression level, and we use DESeq2 based on negative binomial distribution for differential analysis.
Preselected miRNA: After high-throughput sequencing of 8 RNA samples in the OSAS group and the control group, miRNAs with the greatest differences were screened as preselected markers (p value<0.01 and | log2 (foldchange)|>1).
qRT-PCR Validation
To further verify the sequencing results, we performed qRT-PCR on the remaining samples from the two groups (51 in the OSAS group and 52 in the control group). First, RNA reverse transcription was performed on the sample. The Mastercycler PCR instrument of Eppendorf Company was used, and the one-step miRNA reverse transcription kit (Xinhai Gene, D1801) was used. qRT-PCR was performed using the LightCycler 96 fluorescence PCR instrument of ROCHE Company, and 5*Fast SYBR was used. Green qPCR Mix reagent (Xinhai Gene, A2202B). And add the most reliable endogenous reference substance U6 for standardization. Results were normalized using the mean center normalization method, the gold standard method when screening large numbers of miRNAs.
Bioinformatic Analysis
To explore the correlation of miRNAs in the experiments with OSAS, and we further deciphered the potential functions of the identified miRNAs through the mirWalk (http://mirwalk.umm.uni-heidelberg.de/) database. MiRWalk, as a miRNAs-gene interaction prediction database, will be used for target gene prediction among candidate miRNAs. After obtaining the predicted target genes, Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed in the DAVID database.
Statistical Analysis
Mann–Whitney U-test or Student’s T-test will be used to verify the statistical significance of miRNA expression levels between patients and controls. With logistic regression, a combined miRNA panel was built whose profiles included receiver operating characteristic (ROC) curve, area under the ROC curve (AUC), and Youden index (sensitivity + specificity - 1). All data analysis and management work were performed using SPSS 20.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8.3 (GraphPad Software, USA). Data were removed from extreme values and null values, and were stratified by gender.
Results
Ethical Statement
This study was approved by the Ethics Committee of Peking University Shenzhen Hospital and all the participants signed an informed consent form. And this study complied with the Declaration of Helsinki. Totally of 119 participants, including 59 OSAS patients and 60 controls, were enrolled in our study. The study process was shown in Figure 1. The demographics and clinical characteristics of the subjects in our study are shown in Table 1.
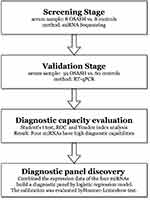 |
Figure 1 The flowchart of the study design. |
To Discover Candidate miRNAs in the Screening Stage
Initially to identify candidate miRNAs in the screening stage, the expression level of miRNAs was screened out with the serums of 8 OSAS patients and 8 controls by miRNA sequencing. Under the standard of p-value<0.01 and | log2 (foldchange)|>1, the candidate miRNAs were chosen based on the differential expression between OSAS patients and controls. Then, 10 miRNAs in serum were most differentially expressed between OSAS patients and controls, including 5 significantly down-regulated miRNAs and 5 significantly up-regulated miRNAs (Figure 2). Thus, these 10 miRNAs were chosen as candidate miRNAs for next study in the validation stage.
To Confirm Candidate miRNAs in the Validation Stage
The expression level of these 10 miRNAs in serum were further assessed by RT-qPCR with 59 OSAS patients and 60 controls, to confirm the expression of these candidate miRNAs serving as serum biomarkers in OSAS diagnosis. As shown in Figure 3, compared to healthy controls, the relative expression level of miR-486-5p, and miR-148a-3p were significantly downregulated in the serum of OSAS patients, and the relative expression level of miR-744-5p, let-7d-3p, and miR-361-3p were significantly increased in the serum of OSAS patients. The other 5 miRNAs, including miR-215-5p, miR-191-5p, miR-182-5p, miR-150-5p, and miR-223-3p, were no difference between OSAS patients and controls.
Diagnostic Value of Candidate miRNAs in the Validation Stage
The diagnostic value of these 10 candidate miRNAs was assessed by ROC curve analysis. The AUCs were 0.745 (95% confidence interval (CI): 0.656 to 0.821; Figure 4A) for miR-486-5p, 0.645 (95% CI: 0.552 to 0.730; Figure 4B) for miR-215-5p, 0.804 (95% CI: 0.721 to 0.871; Figure 4C) for miR-148a-3p, 0.513 (95% CI: 0.418 to 0.607; Figure 4D) for miR-191-5p, 0.565 (95% CI: 0.471 to 0.656; Figure 4E) for miR-182-5p, 0.5 (95% CI: 0.407 to 0.593; Figure 4F) for miR-150-5p, 0.729 (95% CI: 0.639 to 0.807; Figure 4G) for miR-744-5p, 0.525 (95% CI: 0.431 to 0.617; Figure 4H) for miR-223-3p, 0.742 (95% CI: 0.653 to 0.818; Figure 4I) for let-7d-3p, and 0.646 (95% CI: 0.553 to 0.731; Figure 4J) for miR-361-3p, respectively. Youden index was performed to calculate optimum cutoff values and the best sensitivity and specificity of 10 candidate miRNAs in diagnosing OSAS are listed in Table 2.
 |
Table 2 Outcomes of Receiver Operating Characteristic Curves and Youden Index for the ten Candidate miRNAs and the Diagnostic Panel |
Building miRNA Panels for Better Detection of OSAS
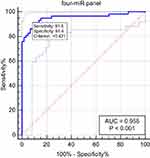
Although, a single miRNA may have good diagnostic values in distinguishing OSAS patients from healthy controls, the combination of several miRNAs could improve higher diagnostic values than single miRNA. By stepwise logistic regression model, the expression data of the miRNAs in the validation stage was combined to build the diagnostic panels. A four-miRNA panel was identified to predict OSAS better and the model was calculated with the formula: Logit (P)= 0.77–1.65 × miR-486-5p - 4.56 × miR-148a-3p + 1.79 × miR-744-5p + 1.13 × let-7d-3p. The AUC for the four-miRNA panel was 0.955 (95% CI: 0.899 to 0.985; sensitivity = 91.38%, specificity = 91.38%; Figure 5).
Bioinformatics Analysis of Candidate miRNAs
Targeted genes prediction was performed by miRWalk database and genes targeted by more than three miRNAs were selected as targeted genes. A total of 367 genes targeted by miR-486-5p, miR-148a-3p, miR-744-5p and let-7d-3p were put into Enrichr database to for KEGG pathway enrichment analysis and GO functional annotation, including biological process (BP), cellular component (CC), and molecular function (MF). The top 10 enriched GO terms in each GO items were shown in Figure 6C, including protein phosphorylation (GO:0006468), RNA interference (GO:0016246), and post-transcriptional gene silencing by RNA (GO:0035194) in the BP category; dendrite (GO:0030425), neuron projection (GO:0043005), and asymmetric synapse (GO:0032279) in the CC category; protein serine/threonine kinase activity (GO:0004674), voltage-gated cation channel activity (GO:0022843), and cytokine receptor binding (GO:0005126) in the MF category. FoxO signaling pathway, Spinocerebellar ataxia, and NF-kappa B signaling pathway were enriched by targeted genes for KEGG analysis (Figure 6D).
Discussion
Obstructive sleep apnea syndrome (OSAS) is a common chronic sleep disorder and influences cardiovascular disease. And previous studies have shown that circulating microRNA could help diagnose OSAS more accurately.9,10 In our study, a two-stage study was designed to identify serum miRNAs that may have diagnostic ability. In the screening stage, the expression level of miRNAs was screened out with the serums of 8 OSAS patients and 8 controls by miRNA sequencing. Then, the most differentially expressed miRNAs were selected as candidate miRNAs for further verification with 59 OSAS patients and 60 controls in the validation stage. We found miR-486-5p, miR-148a-3p, miR-744-5p, and let-7d-3p were significantly dysregulated between OSAS patients and controls. And a four-miRNA panel was identified to detect OSAS, including miR-486-5p, miR-148a-3p, miR-744-5p and let-7d-3p (AUC =0.955 95% CI: 0.899 to 0.985; sensitivity = 91.38%, specificity = 91.38%; Figure 5).
We found miR-486-5p was significantly downregulated in the serum of OSAS patients, and was a good serum biomarker for screening OSAS with AUC=0.745 (95% CI: 0.656 to 0.821; Figure 4A). Also, miR-486-5p has been reported to related to the discrimination between OSAS and non-OSAS patients.11 OSAS was a common disease related to the development of cardiovascular disease, and miR-486-5p in patients with OSAS was associated with the cardiovascular risk SCORE.12,13 The first-choice treatment for sleep apnea is continuous positive airway pressure (CPAP) and miR-486-5p is involved in the favorable response to CPAP treatment in patients with OSA and resistant hypertension through actions on cardiovascular-associated genes and functions.14,15
The miR-148a-3p could be performed to distinguish OSAS patients from controls, among the candidate miRNAs in our study (AUC=0.804; 95% CI: 0.721 to 0.871; Figure 4C), and miR-744-5p (AUC=0.729; 95% CI: 0.639 to 0.807; Figure 4G) and let-7d-3p (AUC=0.742; 95% CI: 0.653 to 0.818; Figure 4I) were also good serum biomarkers for screening OSAS. Previous studies have reported that OSAS is associated with lung cancer.16–22 OSAS patients had higher risk of lung cancer compared with those without OSAS.22 Intermittent nocturnal hypoxemia and sleep fragmentation were immediate consequences of OSAS, which might play a role in carcinogenesis.18–20 And animal studies have shown that OSAS and lung cancer are linked via chronic and intermittent hypoxemia.16,17 And, severe OSAS was associated with increased cancer mortality in patients with stage III–IV lung cancer.21 Studied shown that miR-148a-3p and miR-744-5p were play an important role in the development of lung cancer. Though modulating Ras/MAPK/Erk signaling, miR-148a-3p restrained the epithelial-mesenchymal transition progression and proliferation of non-small-cell lung cancer (NSCLC).23 And miR-148a-3p could regulate Y-box binding protein 1 (YBX1) to promote NSCLC progression.24 Through miR-148a-3p/ DNA methyltransferase 1 (DNMT1) axis, long non-coding RNA HOXA11-AS facilitated NSCLC tumorigenesis in vitro and in vivo.25 By miR-744-5p/ cell division cycle-associated protein 4 (CDCA4) axis, long non-coding RNA LINC01116 promoted lung adenocarcinoma oncogenicity activated by EGR1.26 And, lncRNA MAFG-AS1, served as a molecular sponge of miR-744-5p, boosts the proliferation of lung adenocarcinoma by regulating miR-744-5p/MAFG axis.27 Our study is the first to reveal the role miR-148a-3p, miR-744-5p, and let-7d-3p in the screening of OSAS.
Though KEGG analysis, NF-kappa B signaling pathway were enriched by targeted genes predicted by miR-486-5p, miR-148a-3p, miR-744-5p and let-7d-3p. Obstructive sleep apnea could cause excessive oxidative stress and systemic inflammation by tissue hypoxia. And, reactive oxygen species, induced by hypoxia, affected myoblast pyroptosis though the NF- κ B/HIF-1 α signaling pathway during OSAS.28 Some hypoxia-inducible miRNAs have been identified that be activated in different types of cancers and performing their role in tumorigenesis.29 Oxidative stress and Inflammation significantly correlate with OSAS, DNA oxidation products, TNF-α, NO, IL-6, IL-8, and nitrosative stress compounds could be predictive laboratory biomarkers for OSAS.30–34 OSAS leaded to chronic intermittent hypoxia (CIH) during sleep and was associated with nonalcoholic fatty liver disease, then CIH promoted liver fibrosis by TLR4/MyD88/MAPK/NF-kB signaling pathways in mice.35–37 OSAS was related to cardiovascular morbidity, and NFκB played a role in the development of cardiovascular morbidity in OSAS.37–39 Thus, NF-kappa B signaling pathway played an important role in OSAS.
Conclusion
In conclusion, our study showed that 5 miRNAs (miR-486-5p, miR-148a-3p, miR-744-5p, let-7d-3p, and miR-361-3p) in serum were significantly dysregulated between OSAS patients and healthy controls. The four-miRNA panel (miR-486-5p, miR-148a-3p, miR-744-5p and let-7d-3p) was identified to enhance diagnostic value of screening OSAS, which might be severed as novel biomarker for OSAS diagnosis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sanchez-de-la-Torre M, Khalyfa A, Sanchez-de-la-Torre A, et al. Precision medicine in patients with resistant hypertension and obstructive sleep apnea: blood pressure response to continuous positive airway pressure treatment. J Am Coll Cardiol. 2015;66(9):1023–1032. doi:10.1016/j.jacc.2015.06.1315
2. Tondo P, Fanfulla F, Sabato R, Scioscia G, Foschino Barbaro MP, Lacedonia D. Obstructive Sleep Apnoea-Hypopnoea Syndrome (OSAHS): state of the art. Minerva Med. 2022. doi:10.23736/S0026-4806.22.08190-3
3. Aurora RN, Zak RS, Karippot A, et al. Practice parameters for the respiratory indications for polysomnography in children. Sleep. 2011;34(3):379–388. doi:10.1093/sleep/34.3.379
4. Wu CR, Tu YK, Chuang LP, Gordon C, Chiu HYJSMR. Diagnostic meta-analysis of the pediatric sleep questionnaire, OSA-18, and pulse oximetry in detecting pediatric obstructive sleep apnea syndrome. Sleep Med Rev. 2020;54:101355. doi:10.1016/j.smrv.2020.101355
5. Yang X, Niu X, Xiao Y, Lin K, Chen X. MiRNA expression profiles in healthy OSAHS and OSAHS with arterial hypertension: potential diagnostic and early warning markers. Respir Res. 2018;19(1):194. doi:10.1186/s12931-018-0894-9
6. Li K, Wei P, Qin Y, Wei Y. MicroRNA expression profiling and bioinformatics analysis of dysregulated microRNAs in obstructive sleep apnea patients. Medicine. 2017;96(34):e7917. doi:10.1097/MD.0000000000007917
7. Khalyfa A, Kheirandish-Gozal L, Khalyfa AA, et al. Circulating plasma extracellular microvesicle microrna cargo and endothelial dysfunction in children with obstructive sleep apnea. Am J Respir Crit Care Med. 2016;194(9):1116–1126. doi:10.1164/rccm.201602-0323OC
8. Kapur VK, Auckley DH, Chowdhuri S, et al. Clinical practice guideline for diagnostic testing for adult obstructive sleep apnea: an American Academy of sleep medicine clinical practice guideline. J Clin Sleep Med. 2017;13(3):479–504. doi:10.5664/jcsm.6506
9. Santamaria-Martos F, Benitez I, Ortega F, et al. Circulating microRNA profile as a potential biomarker for obstructive sleep apnea diagnosis. Sci Rep. 2019;9(1):13456. doi:10.1038/s41598-019-49940-1
10. Santamaria-Martos F, Benitez I, Zapater A, et al. Identification and validation of circulating miRNAs as endogenous controls in obstructive sleep apnea. PLoS One. 2019;14(3):e0213622. doi:10.1371/journal.pone.0213622
11. Bradley TD, Floras JS. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009;373(9657):82–93. doi:10.1016/S0140-6736(08)61622-0
12. Santamaria-Martos F, Benitez I, Pinilla L, et al. MicroRNA profile of cardiovascular risk in patients with obstructive sleep apnea. Respiration. 2020;99(12):1122–1128. doi:10.1159/000511093
13. Sanchez-de-la-Torre M, Campos-Rodriguez F, Barbe F. Obstructive sleep apnoea and cardiovascular disease. Lancet Respir Med. 2013;1(1):61–72. doi:10.1016/S2213-2600(12)70051-6
14. Zapater A, Santamaria-Martos F, Targa A, et al. Canonical pathways associated with blood pressure response to sleep apnea treatment: a post hoc analysis. Respiration. 2021;100(4):298–307. doi:10.1159/000511963
15. Niculescu LS, Simionescu N, Sanda GM, et al. MiR-486 and miR-92a identified in circulating HDL discriminate between stable and vulnerable coronary artery disease patients. PLoS One. 2015;10(10):e0140958. doi:10.1371/journal.pone.0140958
16. Almendros I, Montserrat JM, Torres M, et al. Intermittent hypoxia increases melanoma metastasis to the lung in a mouse model of sleep apnea. Respir Physiol Neurobiol. 2013;186(3):303–307. doi:10.1016/j.resp.2013.03.001
17. Almendros I, Montserrat JM, Ramirez J, et al. Intermittent hypoxia enhances cancer progression in a mouse model of sleep apnoea. Eur Respir J. 2012;39(1):215–217. doi:10.1183/09031936.00185110
18. Cabezas E, Perez-Warnisher MT, Troncoso MF, et al. Sleep disordered breathing is highly prevalent in patients with lung cancer: results of the sleep apnea in lung cancer study. Respiration. 2019;97(2):119–124. doi:10.1159/000492273
19. Toffoli S, Michiels C. Intermittent hypoxia is a key regulator of cancer cell and endothelial cell interplay in tumours. FEBS J. 2008;275(12):2991–3002. doi:10.1111/j.1742-4658.2008.06454.x
20. Gozal D, Farre R, Nieto FJ. Putative links between sleep apnea and cancer: from hypotheses to evolving evidence. Chest. 2015;148(5):1140–1147. doi:10.1378/chest.15-0634
21. Huang HY, Lin SW, Chuang LP, et al. Severe OSA associated with higher risk of mortality in stage III and IV lung cancer. J Clin Sleep Med. 2020;16(7):1091–1098. doi:10.5664/jcsm.8432
22. Cheong AJY, Tan BKJ, Teo YH, et al. Obstructive sleep apnea and lung cancer: a systematic review and meta-analysis. Ann Am Thorac Soc. 2022;19(3):469–475. doi:10.1513/AnnalsATS.202108-960OC
23. Xie Q, Yu Z, Lu Y, Fan J, Ni Y, Ma L. microRNA-148a-3p inhibited the proliferation and epithelial-mesenchymal transition progression of non-small-cell lung cancer via modulating Ras/MAPK/Erk signaling. J Cell Physiol. 2019;234(8):12786–12799. doi:10.1002/jcp.27899
24. Su H, Fan G, Huang J, Qiu X. YBX1 regulated by Runx3-miR-148a-3p axis facilitates non-small-cell lung cancer progression. Cell Signal. 2021;85:110049. doi:10.1016/j.cellsig.2021.110049
25. Bai Y, Lang L, Zhao W, Niu R. Long non-coding RNA HOXA11-AS promotes non-small cell lung cancer tumorigenesis through microRNA-148a-3p/DNMT1 regulatory axis. Onco Targets Ther. 2019;12:11195–11206. doi:10.2147/OTT.S198367
26. Ren P, Chang L, Hong X, Xing L, Zhang H. Long non-coding RNA LINC01116 is activated by EGR1 and facilitates lung adenocarcinoma oncogenicity via targeting miR-744-5p/CDCA4 axis. Cancer Cell Int. 2021;21(1):292. doi:10.1186/s12935-021-01994-w
27. Sui Y, Lin G, Zheng Y, Huang W. LncRNA MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR-744-5p/MAFG axis. Eur J Pharmacol. 2019;859:172465. doi:10.1016/j.ejphar.2019.172465
28. Yu LM, Zhang WH, Han XX, et al. Hypoxia-induced ROS contribute to myoblast pyroptosis during obstructive sleep apnea via the NF-kappaB/HIF-1alpha signaling pathway. Oxid Med Cell Longev. 2019;2019:4596368. doi:10.1155/2019/4596368
29. Moriondo G, Scioscia G, Soccio P, et al. Effect of hypoxia-induced micro-RNAs expression on oncogenesis. Int J Mol Sci. 2022;23(11):6294. doi:10.3390/ijms23116294
30. Orru G, Storari M, Scano A, Piras V, Taibi R, Viscuso D. Obstructive sleep apnea, oxidative stress, inflammation and endothelial dysfunction-an overview of predictive laboratory biomarkers. Eur Rev Med Pharmacol Sci. 2020;24(12):6939–6948. doi:10.26355/eurrev_202006_21685
31. Zhang D, Xiao Y, Luo J, et al. Measurement of fractional exhaled nitric oxide and nasal nitric oxide in male patients with obstructive sleep apnea. Sleep Breath. 2019;23(3):785–793. doi:10.1007/s11325-018-1760-1
32. Ip MS, Lam B, Chan LY, et al. Circulating nitric oxide is suppressed in obstructive sleep apnea and is reversed by nasal continuous positive airway pressure. Am J Respir Crit Care Med. 2000;162(6):2166–2171. doi:10.1164/ajrccm.162.6.2002126
33. Oyama J, Yamamoto H, Maeda T, Ito A, Node K, Makino N. Continuous positive airway pressure therapy improves vascular dysfunction and decreases oxidative stress in patients with the metabolic syndrome and obstructive sleep apnea syndrome. Clin Cardiol. 2012;35(4):231–236. doi:10.1002/clc.21010
34. Chami HA, Fontes JD, Vasan RS, et al. Vascular inflammation and sleep disordered breathing in a community-based cohort. Sleep. 2013;36(5):763–768C. doi:10.5665/sleep.2644
35. Kang HH, Kim IK, Lee HI, et al. Chronic intermittent hypoxia induces liver fibrosis in mice with diet-induced obesity via TLR4/MyD88/MAPK/NF-kB signaling pathways. Biochem Biophys Res Commun. 2017;490(2):349–355. doi:10.1016/j.bbrc.2017.06.047
36. Arnaud C, Dematteis M, Pepin JL, Baguet JP, Levy P. Obstructive sleep apnea, immuno-inflammation, and atherosclerosis. Semin Immunopathol. 2009;31(1):113–125. doi:10.1007/s00281-009-0148-5
37. Young T, Shahar E, Nieto FJ, et al. Predictors of sleep-disordered breathing in community-dwelling adults: the sleep heart health study. Arch Intern Med. 2002;162(8):893–900. doi:10.1001/archinte.162.8.893
38. Amin RS, Kimball TR, Bean JA, et al. Left ventricular hypertrophy and abnormal ventricular geometry in children and adolescents with obstructive sleep apnea. Am J Respir Crit Care Med. 2002;165(10):1395–1399. doi:10.1164/rccm.2105118
39. Tal A, Leiberman A, Margulis G, Sofer S. Ventricular dysfunction in children with obstructive sleep apnea: radionuclide assessment. Pediatr Pulmonol. 1988;4(3):139–143. doi:10.1002/ppul.1950040304
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.