Back to Journals » Cancer Management and Research » Volume 10
MALAT1: a potential biomarker in cancer
Authors Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G , Zhu YS
Received 28 March 2018
Accepted for publication 25 June 2018
Published 6 December 2018 Volume 2018:10 Pages 6757—6768
DOI https://doi.org/10.2147/CMAR.S169406
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Harikrishna Nakshatri
Zhi-Xing Li,1,2 Qiong-Ni Zhu,1,2 Hai-Bo Zhang,1,2 Yang Hu,1,2 Guo Wang,1,2 Yuan-Shan Zhu3
1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha, 410008, People’s Republic of China; 2Hunan Key Laboratory of Pharmacogenetics, Institute of Clinical Pharmacology, Central South University, Changsha, 410078, People’s Republic of China; 3Department of Medicine, Weill Cornell Medicine, New York, NY, 10065, USA
Purpose: The research of long non-coding RNAs (lncRNAs) has become a new passion with the discovery of abundant new lncRNAs and extensive investigation of their roles in various diseases, especially in cancers. Metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) emerges as a hotspot, which has been reported to be involved in dysregulation of cell signaling and closely correlated with cancer development, progression, and response to therapy. This review is a brief update of the current knowledge related to the role of MALAT1 in cancer-associated molecular pathways and pathophysiology and possible determinants for MALAT1 to function as a biomarker, aiming to stimulate the basic investigation of lncRNA MALAT1 as well as its translation to clinical applications.
Methods: We have selected vast literature from electronic databases including studies associated with its clinical significance and the pivotal functions in cancer processes such as cell proliferation, apoptosis, metastasis, immunity, angiogenesis, and drug resistance.
Results: Studies have shown that aberrant expression of MALAT1 is related to cancer pathophysiology with the potential to be translated clinically and MALAT1 can regulate cancer processes by interacting with molecules, such as proteins, RNAs and DNAs, and further altering different signal pathways.
Conclusion: MALAT1 lncRNA promises to be a potential biomarker for cancer diagnosis as well as prognosis. Additionally, it might be a therapeutic target for human cancers.
Keywords: MALAT1, lncRNA, cancer therapy, signal pathways, biomarker, chemoresistance, metastasis
Introduction
With the rapid development of high-throughput platforms, the transcriptional landscape of the mammalian genome has been demonstrated. Almost 80% of the genome is transcribed into RNA, while <2% of the genome accounts for protein-coding mRNA.1 Remarkably, a large part of RNAs are non-coding RNAs (ncRNAs), which were once considered transcriptional noise without any biological function. However, their complex and diverse functions have been gradually identified with the advancement of powerful technologies.
NcRNAs can be categorized into two groups according to their length: small ncRNAs and long ncRNAs. In recent years, long ncRNAs that have >200 nucleotides in length are becoming hotspots, and expanding evidence has indicated their complex roles in the regulation of vital cellular function.2 XIST, TSIX, H19, AIR, HOTAIR, NRON, and metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) provide examples of some well-characterized long non-coding RNAs (lncRNAs), and they exert distinctive biological functions through diverse molecular mechanisms. XIST and TSIX mainly function in X-chromosome inactivation.3,4 H19 and AIR are imprinted genes, and H19 is recently reported to have a dual function as an oncogene and a tumor-suppressor gene.5–7 HOTAIR participates in transregulation of gene expression and is associated with tumor invasion and metastasis.8–10 NRON is involved in nuclear trafficking of NFAT.11 Among all these prevalent lncRNAs, MALAT1 has attracted a lot of attention over the past few years, and remarkable progress about MALAT1 has been achieved.
MALAT1, an 8.5 kb lncRNA, located at 11q13, was characterized by Ji et al12 in a study of early-stage non-small-cell lung cancer (NSCLC). MALAT1 (aka α gene) is named due to its clinical significance in predicting metastasis and survival in early-stage NSCLC. A subsequent research study has demonstrated that MALAT1 is widely expressed in normal tissues and conserved among other mammalian species, hinting at a potentially important function in development and evolution.13
Recent research studies have shown that MALAT1 contributes greatly to cancer development and progression. MALAT1 is involved in the modulation of several molecular signaling pathways such as MAPK/ERK,14,15 PI3K/AKT,16 WNT/β-catenin,17 and NF-kB,18 leading to a modification of proliferation, cell death, cell cycle, migration, invasion, immunity, angiogenesis, and tumorigenicity (Figure 1). It is also associated with clinicopathological features including tumor location, tumor size, differentiation, and cancer stage. Furthermore, growing evidence suggests that the aberrant expression of MALAT1 in tumor tissues and/or body fluids may serve as a biomarker for tumor diagnosis and prognosis.
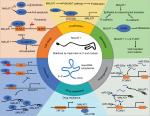  | Figure 1 MALAT1 in cancer pathways. Notes: MALAT1 promotes generation of the six phenotypes of cancer. Diagram partly adapted from Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016; 29(4):452–463).112 Copyright 2016, with permission from Elsevier. Cancer pathways of MALAT1 involved in metastasis, proliferation, cell death, immunity, angiogenesis, and drug resistance are shown. ELAVL1, ELAV-like protein 1 or HuR (human antigen R); NLRP3, NACHT, LRR and PYD domains-containing protein 3; DKK1, Dickkopf1, a Wnt/β-catenin signaling pathway inhibitor; SYK, Spleen tyrosine kinase, also known as Syk; HIF-2a,hypoxia-inducible factor-2a (HIF-2a); FOXM1, Forkhead box protein M1 (FOXM1). Abbreviations: MALAT1, metastasis associated in lung adenocarcinoma transcript 1; SPRR, small proline-rich proteins. |
In the present review, we mainly summarize the recent advancement of MALAT1 related to cancer clinicopathological features and signaling pathways as well as determinants for MALAT1 to be a biomarker in cancer.
MALAT1 involvement in signaling pathways associated with cancer development and progression
Studies over the years have clearly indicated that cancer can generate several pronounced characteristics during its multistep development and progression, including sustaining cell proliferation, resisting cell death, inducing angiogenesis, activating invasion and metastasis, evading immune destruction,19,20 and increasing chemotherapeutic resistance. Recently, accumulating evidence suggests that MALAT1 is one of the important factors to drive these phenotypes by regulating signaling pathways related to the deterioration of cancer. We will therefore briefly describe the specific signaling pathways in which MALAT1 is involved in the regulation of cancer characteristics.
MALAT1 in cell proliferation
LncRNA MALAT1 is generally reported to be an upstream regulator of the MAPK/ERK pathway, which is thought to be one of the most classical pathways referring to cell proliferation.21,22 In retinoblastoma (RB), the MALAT1-induced tumor growth is partly mediated through inhibiting miR-124, then upregulating Slug, a member of MAPK/ERK pathway, leading to an activation of the MAPK/ERK pathway.14 In a Neuro-2a in vitro differentiation cell model, MALAT1 upregulation has been observed and MALAT1 depletion caused a decrease in neurite outgrowth. Furthermore, Neuro-2a cell neurite outgrowth was blocked by PD98059, an inhibitor of ERK/MAPK pathway, whereas an activation of ERK by phorbol 12-myristate 13-acetate rescued the defect in neurite outgrowth due to MALAT1 depletion,15 suggesting that MALAT1 promotes neurite outgrowth through activating the MAPK/ERK pathway. In addition, MALAT1 has been reported to affect cell proliferation through other molecular pathways. In osteosarcoma, both cell proliferation and the phosphorylation of main molecules PI3Kp85α and Akt in the PI3K/AKT signaling pathway were suppressed when MALAT1 expression was downregulated.16 In Ewing’s sarcoma (EWS), MALAT1 may serve as a target of c-MYC that induces MALAT1 expression and cell proliferation.23
MALAT1 in cell death
The regulation of cell death is mediated by many patterns, including apoptosis, autophagy, pyroptosis, and so on. Studies over the years have reported that MALAT1 participated in cell death in a variety of cancers. In multiple myeloma, knockdown of MALAT1 induced apoptosis, which was likely to be associated with the activation of mitochondrial-controlled apoptosis through an upregulation of Bax, Caspase-3/-9 expression and a downregulation of Bcl-2.24 On the other hand, overexpression of MALAT1 inhibited oral tongue squamous cell apoptosis by inducing nuclear β-catenin, whereas the apoptosis is slightly increased when using the Wnt/β-catenin signaling pathway inhibitor DKK1 simultaneously.17 In hepatocellular carcinoma cells (HCC), knockdown of MALAT1 using small-interfering RNAs (siRNAs) inhibited autophagy through inhibiting p62 degradation and puncta formation.25 Additionally, when MALAT1 was repressed in diabetic nephropathy, the expression of ELAVL1, NLRP3, Caspase-1, and the pro-inflammatory cytokine IL-1β, markers of pyroptosis, were downregulated simultaneously,26 indicating that MALAT1 may participate in pyroptosis.
MALAT1 in cancer metastasis
Metastasis is a typical cause of cancer-associated death. It has been documented that the role of MALAT1 in cancer metastasis is primarily modulated by regulating epithelial-to-mesenchymal transition (EMT), a pro-process of metastasis.19,20,27 In ovarian tumor, Jin et al28 have reported that MALAT1 may function as an oncogene, and an inhibition of MALAT1 has significantly impeded EMT with a decrease in matrix metalloproteinase (MMP) and mesenchymal cell markers, including N-cadherin, vimentin, and snail, and an increase in epithelial cell marker, E-cadherin. Furthermore, the PI3K/AKT signaling pathway is verified to be involved in MALAT1 knockdown-mediated reduction of EMT in ovarian cancer cells. In cervical cancer, the expression of MALAT1 was significantly increased in cancer cells and tissues, and the knockdown of MALAT1 in cancer cells inhibited the invasion and metastasis in vitro and in vivo, which may be mediated via attenuating EMT as the epithelial markers, E-cadherin and ZO-1, were upregulated and the mesenchymal markers, β-catenin and vimentin, were downregulated in the MALAT1-knocked-down cells.29 Apart from EMT, MALAT1 has also been reported to promote metastasis through other mechanisms. For example, in tongue cancer, small proline-rich proteins (SPRR) may be involved in MALAT1-mediated tumor metastasis since MALAT1 knockdown causes an upregulation of certain SPRR that influence the distant metastasis of tongue cancer cells.30
MALAT1 in cancer angiogenesis
Basal endothelial sprouting is an indicator of angiogenesis and proliferation.31 It has been reported that MALAT1 regulates angiogenesis through modulating genes associated with cell cycle in endothelial cells.32 In various kinds of endothelial cells, an inhibition of MALAT1 expression resulted in an increase in both endothelial sprout length and the expression of p21 and p27Kip1, genes associated with the inhibition of cell cycle, while a decrease in the number of S-phase cells and in the expression of cyclins: CCNA2, CCNB1, and CCNB2.33 Recently, Li et al34 have demonstrated that MALAT1 plays an important role in vasculogenic mimicry (VM) and angiogenesis in gastric cancer, and they concluded that MALAT1 can promote tumorigenicity and metastasis in gastric cancer cells by facilitating VM and angiogenesis via the VE-cadherin/β-catenin complex and ERK/MMP and FAK/paxillin signaling pathways based on genetic and pharmacological analyses. Moreover, in human umbilical vein endothelial cells, MALAT1 has been shown to competitively bind to miR-320a, partly block the direct interaction between miR-320a and FOXM1, and lead to endothelial cell proliferation and angiogenesis, suggesting that lncRNA MALAT1 may function as a competing endogenous RNA (ceRNA) to promote angiogenesis.35
MALAT1 in cancer immunity
It is now well documented that all tumor lesions have the infiltration of immune cells,36 which possess significant implications in tumorigenicity, tumor progression, and therapy. Recently, many studies have explored the relationship between lncRNA MALAT1 and tumor immunity.18,36,37 In lipopolysaccharide-activated macrophages, MALAT1 was found to interact with NF-kB in the nucleus, leading to an inhibition of NF-kB DNA binding to inflammatory cytokine gene promoters and consequently resulting in a decrease in the production of TNF-α and IL-6.18 In thyroid cancer, it has been shown that the expression of both MALAT1 and FGF2 were enhanced in the tumor tissues and tumor-associated macrophage (TAMs) and that the MALAT1-mediated FGF2 protein secretion from TAMs inhibited inflammatory cytokine release, promoted tumor cell proliferation, migration, and invasion, and induced vasculature formation.37 Taken together, these data suggest that MALAT1 may be involved in tumor immunity mainly through the modulation of tumor microenvironment, especially cytokine production.
MALAT1 in chemotherapeutics resistance
MALAT1 has been reported to contribute to the chemoresistance and radiation resistance of cancer (Table 1). In HCC, MALAT1 upregulation is associated with multiple drug resistance, mediated through an HIF-2α-MALAT1-miR-216b pathway, as evidenced by the findings that the sensitivity of drug-resistant cells to multiple chemotherapeutic agents, including 5-fluorouracil, adriamycin, and mitomycin C, was elevated when MALAT1 siRNA and miR-216b mimics were used.25 In studies investigating the resistance of glioblastoma to temozolomide (TMZ), MALAT1 has been shown to modify drug sensitivity through the regulation of MALAT1/ZEB1/MDR pathway as well as the expression of thymidylate synthase (TS) and miR203; besides, the induced miR-101 expression by MALAT1 knockdown can reverse the chemoresistance to TMZ.38–40 Similarly, in colorectal cancer, MALAT1 overexpression is associated with oxaliplatin resistance and poor patient survival.41 Furthermore, in lung cancer, Wang et al42 have found that MALAT1 knockdown in cisplatin-resistant A549/DDP cells resulted in miR-101-3p upregulation and MCL1 downregulation, which consequently increased cisplatin sensitivity; moreover, the polymorphism, rs619586, of MALAT1 has been demonstrated to be notably associated with platinum-based chemotherapeutic response, which may serve as a potential biomarker for the selection of platinum-based chemotherapy response in lung cancer patients.43 In addition, MALAT1 has been reported to be involved in radiotherapy resistance of nasopharyngeal carcinoma, in which lncRNA MALAT1 was upregulated in tumor tissues and knockdown of MALAT1 potentiated cell sensitivity to radiotherapy both in vitro and in vivo.44 These data collectively indicate that MALAT1 may serve as a potential therapeutic target in multiple cancers.
  | Table 1 MALAT1 and drug resistance Abbreviations: ADR, adriamycin; 5-FU, 5-fluorouracil; MALAT1, metastasis associated in lung adenocarcinoma transcript 1; MMC, mitomycin C; TMZ, temozolomide. |
Generally, the hallmarks generated during cancer development and progression are caused by mutations that produce oncogenes and tumor suppressor genes, and the altered expression of these genes can greatly influence fundamental biological processes, such as cell proliferation, cell death, cell metastasis, and so on. The oncogenes and tumor suppressor genes with similar functions can generate a signaling pathway, involving the regulation of multiple biological processes. MALAT1 can interact with these oncogenes and tumor suppressors, thus participating in various signaling pathways and regulating the processes associated with cancer development and progression. Currently, the research studies of MALAT1 mainly focus on cancer metastasis, an indicator of cancer deterioration, which is possibly due to MALAT1 involvement in EMT and the closed correlation with lymph node metastasis. These research studies in cancer collectively indicate that MALAT1 may be an anchor which bridges multiple pathological processes associated with cancer, and it can be a potential biomarker for cancer.
Mechanisms of MALAT1 function
Recent studies have documented that MALAT1 represents a functional molecule that displays vital roles not only in clinical but also in diverse physiological and pathological processes,1,45,46 such as cell proliferation, cell death, cell cycle, cell migration, invasion, immunity, and angiogenesis via involvement of signaling pathways, making it a possible biomarker and drug target. However, this also raises several questions: how does MALAT1 fulfill its function mechanistically? Human body is a huge factory, which produces millions of proteins, RNAs, and DNAs, and the functions of these molecules are important for its biological processes.
Many studies over the years have revealed that lncRNAs fulfill their functions through directly or indirectly interacting with other molecules such as proteins, RNAs, or DNAs.47 Although the molecular mechanisms of MALAT1 actions are not fully understood, we will highlight some of the mechanisms associated with MALAT1 actions in diverse cellular processes.
MALAT1 interaction with protein
To date, many proteins have been shown to interact with MALAT1. MALAT1 predominantly is retained in nuclear speckles,48 eliciting possible functional binding to the serine/arginine-rich (SR) family splicing factors, which locate at nuclear speckles, as well as functioning as modulators of alternative splicing (AS). Tripathi et al49 conducted a series of computational and biochemical assays and found that MALAT1 can interact with several splicing factors, such as SRSF1 (ASF/SF2), SC35 (SRSF2), and SRSF3. Interestingly, depletion of SR proteins does not change the speckle localization of MALAT1, whereas depletion of MALAT1 influences the speckle distribution and phosphorylation of SR proteins, indicating a functional role of MALAT1 in AS.49 In hepatocellular carcinoma, MALAT1 induces the expression of oncogenic splicing factor SRSF1, which enhances the production of antiapoptotic splicing isoforms and modulates the AS of S6K1 to activate the mTOR pathway, thus promoting the development of hepatocellular carcinoma.50 In breast cancer (BC), mutant p53 and ID4 proteins influence the interaction between MALAT1 and SRSF1 and intranuclear localization of MALAT1 to active transcription sites. Further study shows that mutant p53 and ID4 favor the interaction of MALAT1 with VEGFA precursor mRNA, stabilizing binding of SRSF1 to this precursor and modulating the expression of VEGFA isoforms and finally impacting the angiogenesis of BC cells.51 These results are consistent with the former study that repression of MALAT1 could alter the pattern of AS of particular pre-mRNAs. Recent studies have shown that MALAT1 and TUGA1 play important role in the regulation of proliferation. MALAT1 directly binds to the unmethylated Polycomb 2 (Pc2) on the promoter of E2F1, a critical regulator of cell proliferation, leading to E2F1 SUMOylation and the relocation of the growth control genes from transcriptional repressive Pc2 bodies to a transcriptionally permissive environment of the interchromatin granules. On the contrary, TUGA1 binds to the methylated Pc2, resulting in the growth control gene promoters retaining in Pc2 bodies and thus inhibiting gene expression and cell proliferation.52 A systematic research delineates that MALAT1 competitively binds to depleted in breast cancer 1 (DBC1) from a sirtuin1 (SIRT1)–DBC1 complex, leading to the release of SIRT1. SIRT1 has an increased deacetylation activity and promotes the acetylation of p53, which subsequently impairs P53 function in promoting cell apoptosis and inhibiting cell growth.53 Similarly, in colorectal cancer, MALAT1 promotes tumor growth and metastasis by competitively binding to SFPQ, a tumor suppressor, thus releasing proto-oncogene PTBP2 from the SFPQ/PTBP2 complex. Studies conducted by Yang et al54 have shown that the interaction between MALAT1 and a nuclear protein hnRNP C promotes the translocation of MALAT1 from the nucleus into the cytoplasm, which consequently increases mitosis and promotes the transition of G2/M phase. RNA–protein interaction studies show that MALAT1 can bind to several other proteins as well as affect the pathophysiology of cancer. These data collectively indicate that the interaction of MALAT1 with some RNA-related proteins is critical for the regulation of cellular biological processes.
MALAT1 interaction with RNA
Except for proteins, MALAT1 can interact with RNAs to achieve its diverse molecular functions. Using RAP-RNA, an assay to detect intermolecular RNA–RNA interactions, Engreitz et al55 have observed that Malat1 could interact with nascent transcripts indirectly through protein intermediates, which caused Malat1 to localize proximally to chromatin at active genes, suggesting that Malat1 may influence RNA processing through recruitment or modification of other proteins localized to these sites.
The importance of ceRNA network in cancer pathophysiology has become evident in recent years. MicroRNAs are important regulators in the ceRNA network that negatively regulates gene expression of mRNAs. The most commonly observed mechanism is that microRNAs interact with mRNA 3′-UTRs of target gene, leading to deadenylation, decreased mRNA stability, and translation suppression.56 Recent evidence has shown that other RNAs can compete with mRNAs for shared microRNAs.57 Among these RNAs, MALAT1 is one of the most studied as a ceRNA to be involved in various molecular processes (Table 2).
The strongest evidence of MALAT1 as a ceRNA comes from studies on cancer. In BC, MALAT1 and cell division cycle 42 (CDC42) compete for the binding of miR-1, consequently reducing CDC42 expression and leading to increased ability of migration and invasion.58 Another study showed that MALAT1 regulates the EMT of BC through competitively binding to miR-204 with ZEB2, a target gene of miR-204.59 In osteosarcoma, upregulation of MALAT1 expression is observed in both human osteosarcoma cell lines and tissues, which promotes osteosarcoma cell growth. Mechanical research studies showed that MIR376A is shared by TGFA and MALAT1, and upregulation of MALAT1 results in decreased binding of MIR376A to TGFA, thus increasing TGFA expression and osteosarcoma cell growth.60 In gastric cancer, the MALAT1/miR-202/Gli2 regulatory pathway may explain the clinical relevance with tumor size, lymph node metastasis, and TNM stage and the alteration of some molecular processes such as proliferation, cell cycle, and cell apoptosis.61 In hepatocellular carcinoma, the alteration in regulatory network of two ceRNAs, MALAT1-miR-195-EGFR and MALAT1 miRNA-204-SIRT1, is responsible for the increased ability of migration and invasion.62,63 In gallbladder cancer, MALAT1 functions as an oncogene with high expression in tumor tissues and cell lines. Recent studies showed that MALAT1 modifies proliferation, apoptosis, cell cycle, and invasion by acting as a molecular sponge to negatively regulate miR-206 or miR-363-3p, thus resulting in the upregulation of ANXA2 and KRAS or MCL-1.64,65 In glioma, MALAT1 conversely plays a tumor-suppressive function with inhibited cell viability, which is mediated by downregulating miR-155 and upregulating FBXW7 expression. Another study demonstrated that miR-129 can function as a linker, which directly bridges MALAT1 and Sox4, and its expression negatively correlates with the expression of MALAT1 and Sox4, indicating that the disorder of MALAT1/miR-129/Sox4/pathway may account for the enhanced glioma stem cell viability and proliferation abilities and promoted glioma tumorigenesis.66,67
In addition, the abnormal interaction between MALAT1 and micro-interfering RNA (miRNA) can be seen in other cancers, such as ovarian cancer,68,69 cervical cancer,70 and melanoma.71
MALAT1 interaction with DNA
Considering the intrinsic nature of MALAT1, an RNA, it has an easy access to bind to homologous DNA sequence. West et al72 applied Capture Hybridization Analysis of RNA Targets (CHART), a technology developed to identify putative trans genomic binding sites for endogenous RNAs and to map the genomic binding sites for two highly expressed lncRNAs, NEAT1 and MALAT1. Subsequent analyses demonstrated that NEAT1 and MALAT1 can interact with numerous active genes and NEAT1 and MALAT1 can colocalize to many transcribed gene loci while with different binding patterns, indicating a possible mechanism that MALAT1 may modify transcription through binding to actively transcribed gene loci.72 In renal tumor cells, the MALAT1 (aka Alpha) gene locus is found to translocate to that of transcription factor EB (TFEB), leading to an Alpha-TFEB fusion. MALAT1 promoter induces the expression of this fusion gene, actually the upregulation of TFEB due to the fact that MALAT1 does not contribute to the ORF. The upregulation of TFEB transcription consequently results in tumorigenesis.73 We can infer that a mechanism by which MALAT1 regulates transcription is through translocation and fusion of the MALAT1 gene locus to another gene locus.
The clinical significance of MALAT1
Epidemiological study has indicated that cancer is a leading cause of death.74 Most cancers are hard to be cured due to the limited diagnostic and therapeutic methods. Since MALAT1 was first identified to stratify early-stage NSCLC patients at high risk to develop metastasis,12 the later research studies paid more attention to its clinical significance (Table 3).76–83 Herein, we briefly introduce the clinical significance of MALAT1 in commonly diagnosed cancers.
MALAT1 serves as a biomarker for cancer diagnosis
Early cancer detection is crucial for a successful cancer therapy. Although various cancer diagnostic methodologies are available, it is unfortunate that these methods do not fully meet the demand of early cancer diagnosis, and new methods with high specificity and sufficient sensitivity are needed. Studies over the years have indicated that lncRNA MALAT1 may serve as a diagnostic biomarker with sufficient specificity and sensitivity for multiple malignancies (Table 3). The diagnostic value of a biomarker is estimated through the receiver operating characteristic (ROC) curve, which is defined as diagnostic accuracy. The accuracy of tests is based on a rough rule of thumb. If the area under the ROC curve (AUC) is between 0.50 and 0.70, the accuracy is low; if AUCs are over 0.90, the accuracy is high and those between these two are considered moderate.75
In a clinical study of NSCLC with a small sample size, different MALAT1 expression levels in human peripheral blood are observed between cancer patients and cancer-free controls, and ROC analysis has demonstrated that MALAT1 might not be an independent diagnostic biomarker due to its relatively low sensitivity, whereas it can be used with other diagnostic biomarkers to improve the entire sensitivity and specificity.76 However, in a relatively larger clinical trial, the investigators have revealed that the expression of a circulating ncRNA panel consisting of MALAT1, miR-1254, miR-485-5p, and miR-574-5p can distinguish NSCLC patients from controls with sufficient specificity and sensitivity.77 Moreover, serum exosomal MALAT1 level could be used as a diagnostic biomarker for the metastasis of NSCLC.78 In BC patients, the serum MALAT1 level is distinguishably increased compared to benign breast tumor patients, which is fairly satisfactory to serve as a diagnostic biomarker of BC.79 A serum circulating lncRNA panel, including MEG3, SNHG16, and MALAT1, has been identified for bladder cancer diagnosis, which has a significantly higher AUC compared to the urinary cytology, indicating a far superior diagnostic performance.80 Furthermore, MALAT1 has been used for the diagnosis of other types of cancers, such as nasopharyngeal carcinoma,81 osteosarcoma,82 and epithelial ovarian cancer.83. These data collectively indicate that MALATA1 alone or in combination with other molecules may serve as a useful biomarker for the diagnosis of various cancers.
MALAT1 serves as a biomarker for cancer prognosis
Recently, aberrant expression of MALAT1 has been reported to be associated with cancer prognosis markedly (Table 4). In NSCLC, a higher MALAT1 expression significantly related to female sex (P=0.019), TNM stage (P=0.016), vessel invasion (P=0.032), pathological differentiation (P=0.013), and recurrence (P=0.006).84 Additionally, cooverexpression of MALAT1, NEAT1, and Oct4 in lung cancer has been shown to be an independent prognostic factor for poor outcome.85 Further study has indicated that MALAT1 overexpression is significantly related to the prognosis of lung squamous cell carcinoma but not non-squamous cell carcinoma.86 Genetic variants of MALAT1 are associated with survival outcome in advanced lung adenoma patients, such as the rs3200401 T allele, which is a protective factor, and advanced lung adenoma patients with rs3200401 T allele have a longer median survival time (MST) and better prognosis.87 In BC, MALAT1 is overexpressed in cancerous tissues, which is dramatically correlated with lymph metastasis, adverse 5-year disease-free survival and poor relapse-free survival in a cancer subtype-dependent manner.79,88–90 In colorectal cancer, the overexpression of MALAT1 in tumor tissues is related to a relatively shorter survival time.91,92 In nasopharyngeal carcinoma, MALAT1 overexpression is associated with advanced TNM stage and EBV infection, and an alteration of serum MALAT1 is a predictive biomarker for therapeutic outcome.81 In osteosarcoma, patients with higher MALAT1 expression show poor overall survival (OS) and progressive-free survival (PFS) compared to those with lower MALAT1 expression.82 In bladder cancer, overexpression of MALAT1 in tumor tissues is associated with poor OS, metastatic lymph node, advanced histological grade, and higher tumor stage, indicating that MALAT1 may serve as an independent prognostic biomarker for bladder cancer.93 Taken together, these studies provide promising data that MALAT1 may be an independent biomarker for the prediction of various cancer outcomes and warrant further clinical investigations with larger sample sizes.
MALAT1 serves as a target for cancer therapy
Advances in gene therapy will contribute to the treatment of a large number of genetic diseases as well as cancers because of the flexibility, efficiency, and reduced off-target effects of the innovative treatments.94 The current research of MALAT1 in cancer95 and methodological advances in targeting tumor cells have provided MALAT1 an opportunity to serve as a target for cancer therapy. In osteosarcoma, in vitro knockdown of oncogene MALAT1 leads to the inducement of cell cycle arrest and apoptosis, whereas the decrease in cell proliferation and migration. Besides, the xenograft model verified that MALAT1 silencing inhibits tumorigenesis of osteosarcoma cells, which collectively indicates that MALAT1 lncRNA can be targeted for osteosarcoma treatment.96 Studies conducted by Gutschner et al97 have unraveled that lung cancer cells with a 1,000-fold reduction of MALAT1 using Zinc-finger nucleases (ZFNs) showed impaired ability in migration and formed significantly fewer and smaller lung tumor nodules in a xenograft lung cancer mouse model, suggesting that decreasing MALAT1 expression may be a potential therapeutic strategy for metastasizing lung cancer. Additionally, the mouse with luminal B BC can benefit from the defect in Malat1, which facilitates the transformation from aggressive mammary carcinomas with higher possibility of lung metastasis to cystic tumors that are less prone to metastasizing, giving a strong clue on the further clinical strategy of targeting MALAT1 for the treatment of BC.98 Similarly, another independent group has also verified that Malat1 deficiency in the MMTV-PyMT mammary carcinoma mouse model significantly influences AS and the expression of molecules in pro-tumorigenic signaling pathways, thus resulting in a better pathological feature with higher differentiation and less metastasis. These data show that MALAT1 may serve as a target for inducing differentiation of primary human breast tumors and significantly reducing the risk of tumor metastasis.99 Furthermore, Wheeler et al100 have proved that antisense oligonucleotides (ASOs) based on the RNase H-dependent antisense knockdown are efficient to knockdown the nuclear retained RNAs such as lncRNA MALAT1. Short double-stranded RNA molecules and genome-editing techniques, including siRNAs,101 miRNAs,102 ZFNs, transcription activator-like effector nucleases (TALENs) and clustered, regularly interspaced, short palindromic repeat (CRISPR)/CRISPR associated (Cas)-9 systems,103 have emerged as powerful strategies for the treatment of various diseases with potential therapeutic value. Together, these data demonstrate that MALAT1 may represent a candidate therapeutic target, and MALAT1 knockdown strategies may represent potential therapies for inhibiting cancer progression.
Characteristics of MALAT1 as a potential biomarker
Generally, an ideal biomarker should possess several typical characteristics. First, it should be easily obtained with minimal risk and discomfort to patients. Second, it should be measurable and/or quantifiable with high sensitivity and specificity, making it easy to be detectable and reproducible in standard clinical laboratories. In addition, it should be of high specificity and its expression should be altered significantly in cancer patients.104,105
MALAT1 lncRNA possesses all the features as a biomarker. MALAT1 is expressed not only in tissues but also detectable in body fluids such as blood,45 a sample source easily obtainable with minimal risk to the patient. The advancement in technology makes it possible to detect low-abundant RNA transcripts through PCR amplification and RNA-seq in clinical laboratories. It is evident that the differential expression of MALAT1 lncRNA in diverse cancers is present, which is associated with clinical characteristics and drug resistance in cancer patients,38,39 suggesting that MALAT1 lncRNA is a potential biomarker. However, few studies have currently emphasized the cell-type specificity and intratumor heterogeneity of MALAT1 expression in cancer patients, which might be addressed using fluorescent in situ hybridization (ISH or FISH) and single-cell RNA-seq. Furthermore, MALAT1 lncRNA is a relatively stable RNA transcript with a half-life of 9–12 hours, presumably due to its triple helix structure at the 3′-end.106–108 This long half-life feature makes it easily detectable in tumor tissues as well as in body fluids. Regrettably, unlike PCA3 lncRNA, a prostate cancer-specific biomarker approved by the US Food and Drug Administration (FDA) in 2012 for prostate cancer diagnosis,105 MALAT1 is expressed across a broad range of cell types, making it less specific to distinguish the origin of tumors when it is used as a diagnostic biomarker in body fluids. Nevertheless, a hematic detection of MALAT1 may serve as a supplement for the clinical diagnosis of cancer patients to enhance diagnostic sensitivity and accuracy.
Conclusion and prospective
As a prominent lncRNA, MALAT1 has been widely studied in the last several years, especially its roles in cancer development, metastasis, drug resistance, and clinical outcome. However, it is puzzling that knockout of MALAT1 in animals has no robust phenotypes in development, gene expression, and physiological functions.13,109,110 Studies by Spector’s group in Cold Spring Harbor Laboratory have demonstrated in a mouse Malat1 loss-of-function genetic model that lncRNA Malat1 was dispensable for mouse development, but it might play a cis-regulatory role in neighboring gene expression in adulthood.109 This observation is consistent with the findings by other independent groups, who have reported that a loss of MALAT1 was compatible with life and development13 and had no impact on physiological functions.110 Taken together, these observations suggest that MALAT1 per se may contribute moderately to normal development and physiological functions in mammals. However, through interacting with other RNAs, proteins, and DNA,54,55,111 lncRNA MALAT1 may possess significant roles in pathophysiology, especially in cancer development, progression, and response to therapy, presumably mediated via various molecular pathways. In spite of the abovementioned mechanisms, there are still some questions. For example, are there any triggers for the alteration of MALAT1? Why does MALAT1 play different roles in various cell lines? Do the different transcripts of MALAT1 have distinct functions? Elucidation of the molecular pathways and the interacting networks of MALAT1 with other molecules will facilitate the understanding of its roles in the pathogenesis of various diseases and pave the way for in-depth knowledge of MALAT1, leading to clinical applications in the near future.
Acknowledgments
This review was funded by the National Natural Science Foundation of China (number 81673516) and the Special Talents Fund from Central South University of China.
Disclosure
The authors report no conflicts of interest in this work.
References
Yoshimoto R, Mayeda A, Yoshida M SN. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–199. | ||
Gutschner T SD. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. | ||
Lee J, Davidow LS, Warshawsky D, Tsix DW. A gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21(4):400–404. | ||
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. | ||
Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. | ||
Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. | ||
Rinn JL, Kertesz M, Wang JK, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129(7):1311–1323. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Pastori C, Kapranov P, Penas C, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proceedings of the National Academy of Sciences. 2015;112(27):8326–8331. | ||
Willingham AT, Orth AP, Batalov S, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. | ||
Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. | ||
Eißmann M, Gutschner T, Hämmerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. | ||
Liu S, Yan G, Zhang J, Yu L. Knockdown of Long Noncoding RNA (lncRNA) Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Inhibits Proliferation, Migration, and Invasion and Promoted Apoptosis By Targeting miR-124 in Retinoblastoma. Oncol Res. Epub 21 May 2017. | ||
Chen L, Feng P, Zhu X, et al. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J Cell Mol Med. 2016;20(11):2102–2110. | ||
Dong Y, Liang G, Yuan B, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor Biology. 2015;36(3):1477–1486. | ||
Liang J, Liang L, Ouyang K, Li Z, Yi X. MALAT1 induces tongue cancer cells’ EMT and inhibits apoptosis through Wnt/β-catenin signaling pathway. J Oral Pathol Med. 2017;46(2):98–105. | ||
Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016;590(17):2884–2895. | ||
Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100(1):57–70. | ||
Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144(5):646–674. | ||
Mccubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 1773;2007(8):1263–1284. | ||
Han Y, Wu Z, Wu T, et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7(3):e2123. | ||
Sun H, Lin D-C, Cao Q, et al. Identification of a Novel SYK/c-MYC/MALAT1 Signaling Pathway and Its Potential Therapeutic Value in Ewing Sarcoma. Clinical Cancer Research. 2017;23(15):4376–4387. | ||
Liu H, Wang H, Wu B, et al. Down-regulation of long non-coding RNA MALAT1 by RNA interference inhibits proliferation and induces apoptosis in multiple myeloma. Clinical and Experimental Pharmacology and Physiology. 2017;44(10):1032–1041. | ||
Yuan P, Cao W, Zang Q, et al. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478(3):1067–1073. | ||
Li X, Zeng L, Cao C, et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res. 2017;350(2):327–335. | ||
Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331(6024):1559–1564. | ||
Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21(14):3176–3184. | ||
Sun R, Qin C, Jiang B, et al. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial–mesenchymal transition. Mol Biosyst. 2016;12(3):952–962. | ||
Fang Z, Zhang S, Wang Y, et al. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer. 2016;16(1):706. | ||
Hillen F, Griffioen AW. Tumour vascularization: sprouting angiogenesis and beyond. Cancer and Metastasis Reviews. 2007;26(3-4):489–502. | ||
Li X, Song Y, Liu F, et al. Long Non-Coding RNA MALAT1 Promotes Proliferation, Angiogenesis, and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J Cell Biochem. 2017;118(9):2780–2791. | ||
Michalik KM, You X, Manavski Y, et al. Long Noncoding RNA MALAT1 Regulates Endothelial Cell Function and Vessel Growth Novelty and Significance. Circ Res. 2014;114(9):1389–1397. | ||
Li Y, Wu Z, Yuan J, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. | ||
Jy S, Zw Z, Wm L, et al. Knockdown of MALAT1 expression inhibits HUVEC proliferation by upregulation of miR-320a and downregulation of FOXM1 expression. Oncotarget. 2017;8(37):61499–61509. | ||
Finn OJ, Immunology C. N Engl J Med. 2008;358:2704–2715. | ||
Huang J-Kang, Ma L, Song W-Hua, et al. LncRNA-MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor-Associated Macrophage FGF2 Protein Secretion. J Cell Biochem. 2017;118(12):4821–4830. | ||
Li H, Yuan X, Yan D, et al. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cellular Physiology and Biochemistry. 2017;42(3):1192–1201. | ||
C W, Xk X, Jl L, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8(14):22783–22799. | ||
Cai T, Liu Y, Xiao J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018;7(4):1404–1415. | ||
Li P, Zhang X, Wang H, et al. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16(4):739–751. | ||
Wang HWL, Zhang G, Lu C, et al. MALAT1/miR-101-3p/MCL1 axis mediates cisplatin resistance in lung cancer. Oncotarget. 2017;9(7):7501–7512. | ||
Gong WJ, Yin JY, Li XP, et al. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37(6):8349–8358. | ||
Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37(3):4025–4033. | ||
Wei Y, Niu B. Role of MALAT1 as a Prognostic Factor for Survival in Various Cancers: A Systematic Review of the Literature with Meta-Analysis. Dis Markers. 2015;2015(6):164635–164639. | ||
Gutschner T, Hämmerle M, Diederichs S. MALAT1 — a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91(7):791–801. | ||
Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. | ||
Clemson CM, Hutchinson JN, Sara SA, et al. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol Cell. 2009;33(6):717–726. | ||
Tripathi V, Ellis JD, Shen Z, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol Cell. 2010;39(6):925–938. | ||
Malakar P, Shilo A, Mogilevsky A, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77(5):1155–1167. | ||
Pruszko M, Milano E, Forcato M, et al. The mutant p53‐ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017;18(8):1331–1351. | ||
Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 Methylation-Dependent Gene Relocation between Nuclear Structures Mediates Gene Activation Programs. Cell. 2011;147(4):773–788. | ||
Chen R, Liu Y, Zhuang H, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45(17):9947–9959. | ||
Yang F, Yi F, Han X, du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587(19):3175–3181. | ||
Engreitz JM, Sirokman K, Mcdonel P, et al. RNA-RNA Interactions Enable Specific Targeting of Noncoding RNAs to Nascent Pre-mRNAs and Chromatin Sites. Cell. 2014;159(1):188–199. | ||
Cortés-Lópeza M, Miura P. Emerging Functions of Circular RNAs. Yale J Biol Med. 2016;89(4):527–537. | ||
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. | ||
Chou J, Wang B, Zheng T, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472(1):262–269. | ||
Wang Y, Zhou Y, Yang Z, et al. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial–mesenchymal transition in breast cancer. Tumor Biology. 2017;39(7):1010428317690998. | ||
Luo W, He H, Wang Q, et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7(34):54733–54743. | ||
Zhang Y, Chen Z, Li MJ, Guo HY, Jing NC. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. Biomed Pharmacother. 2017;85:264–271. | ||
Liu D, Zhu Y, Pang J, et al. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 2018;119(2):1368–1380. | ||
Hou Z, Xu X, Zhou L, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumor Biology. 2017;39(7):1010428317718135. | ||
Wang SH, Zhang WJ, Wu XC, et al. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 2016;20(12):2299–2308. | ||
Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857–37867. | ||
Cao S, Wang Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am J Cancer Res. 2016;6(11):2561–2574. | ||
Xiong Z, Wang L, Wang Q, Yuan Y. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J Cell Mol Med. 2018;93(12). | ||
Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther. 2017;10:35–46. | ||
Tao F, Tian X, Ruan S, Shen M, Zhang Z. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. Faseb J. Epub 6 Jun 2018. | ||
Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016;37(1):633–640. | ||
Luan W, Li L, Shi Y, et al. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7(39):63901–63912. | ||
West JA, Davis CP, Sunwoo H, et al. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol Cell. 2014;55(5):791–802. | ||
Davis IJ, Hsi B-L, Arroyo JD, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proceedings of the National Academy of Sciences. 2003;100(10):6051–6056. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. | ||
Streiner DL, Cairney J. What’s Under the ROC? An Introduction to Receiver Operating Characteristics Curves. Research Methods in Psychiatry. 2007;52(2):121–128. | ||
Weber D, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6(1):518. | ||
Peng H, Wang J, Li J, et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. | ||
Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–414. | ||
Miao Y, Fan R, Chen L, et al. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Annals of Clinical & Laboratory Science. 2016;46(4):418–424. | ||
Duan W, Du L, Jiang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850–78858. | ||
He B, Zeng J, Chao W, et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8(25):41166–41177. | ||
Huo Y, Li Q, Wang X, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8(29):46993–47006. | ||
Chen Q, Su Y, He X, et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12(2):1361–1366. | ||
Lin L, Li H, Zhu Y, et al. Expression of metastasis-associated lung adenocarcinoma transcript 1 long non-coding RNA in vitro and in patients with non-small cell lung cancer. Oncol Lett. 2018;15(6):9443–9449. | ||
Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16(1):104. | ||
Schmidt LH, Spieker T, Koschmieder S, et al. The Long Noncoding MALAT-1 RNA Indicates a Poor Prognosis in Non-small Cell Lung Cancer and Induces Migration and Tumor Growth. Journal of Thoracic Oncology. 2011;6(12):1984–1992. | ||
Wang JZ, Xiang JJ, Wu LG, et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17(1):167. | ||
Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7(26):40418–40436. | ||
Huang NS, Chi YY, Xue JY, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7(25):37957–37965. | ||
Wang Z, Katsaros D, Biglia N, et al. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res Treat. 2018;171(2):261–271. | ||
Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. | ||
Li Q, Dai Y, Wang F, Hou S. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63(6):977–983. | ||
Li C, Cui Y, Liu L-F, et al. High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin Genitourin Cancer. 2017; 15(5):570–576. | ||
Husain SR, Han J, Au P, Shannon K, Puri RK. Gene therapy for cancer: regulatory considerations for approval. Cancer Gene Ther. 2015;22(12):554–563. | ||
Ren D, Li H, Li R, et al. Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett. 2016;11(3):1621–1630. | ||
Cai X, Liu Y, Yang W, et al. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. Journal of Orthopaedic Research. 2016;34(6):932–941. | ||
Gutschner T, Hämmerle M, Eissmann M, et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013;73(3):1180–1189. | ||
Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. N Engl J Med Overseas Ed. 2016;374(23):2287–2289. | ||
Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. | ||
Wheeler TM, Leger AJ, Pandey SK, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. | ||
Ryther RC, Flynt AS, Phillips JA, Patton JG. siRNA therapeutics: big potential from small RNAs. Gene Ther. 2005;12(1):5–11. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. | ||
Gutschner T, Richtig G, Haemmerle M, Pichler M. From biomarkers to therapeutic targets-the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018;37(1):83–105. | ||
Deng J, Tang J, Wang G, Zhu YS. Long Non-Coding RNA as Potential Biomarker for Prostate Cancer: Is It Making a Difference? Int J Environ Res Public Health. 2017;14(3):270. | ||
Tani H, Nakamura Y, Ijiri K, Akimitsu N. Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov Ther. 2010;4(4):235–239. | ||
Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–2407. | ||
Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109(47):19202–19207. | ||
Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. | ||
Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. | ||
Macias S, Plass M, Stajuda A, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19(8):760–766. | ||
Schmitt AM, Chang HY. Long Nnoncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



