Back to Journals » Drug Design, Development and Therapy » Volume 16
Mahuang Decoction Attenuates Airway Inflammation and Remodeling in Asthma via Suppression of the SP1/FGFR3/PI3K/AKT Axis
Authors Wei L, Gou X, Su B, Han H, Guo T, Liu L, Wang L, Zhang L, Chen W
Received 16 December 2021
Accepted for publication 3 August 2022
Published 26 August 2022 Volume 2022:16 Pages 2833—2850
DOI https://doi.org/10.2147/DDDT.S351264
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Tuo Deng
Lina Wei,1 Xulei Gou,2 Baoning Su,3 Haiqiong Han,4 Tingting Guo,5 Liang Liu,5 Lei Wang,5 Lina Zhang,3 Weibin Chen6
1Children’s Medical Center, Affiliated Hospital of Changchun University of Traditional Chinese Medicine, Changchun, 130021, People’s Republic of China; 2Department of Pediatrics, Beijing Daxing District Maternal and Child Health Care Hospital, Beijing, 102600, People’s Republic of China; 3Department of Pediatrics, Shanghai Jiading Traditional Chinese Medicine Hospital, Shanghai, 201800, People’s Republic of China; 4Department of Rehabilitation Medicine, Community Health Service Center of Jiangqiao Town in Jiading District, Shanghai, 201803, People’s Republic of China; 5Changchun University of Traditional Chinese Medicine, Changchun, 130017, People’s Republic of China; 6Department of Pediatrics, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, 200437, People’s Republic of China
Correspondence: Lina Zhang, Department of Pediatrics, Shanghai Jiading Traditional Chinese Medicine Hospital, No. 222, Bole Road, Jiading District, Shanghai, 201800, People’s Republic of China, Tel +86-18930568760, Email [email protected] Weibin Chen, Department of Pediatrics, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, No. 110, Ganhe Road, Hongkou District, Shanghai, 200437, People’s Republic of China, Tel +86-18621863233, Email [email protected]
Background/Purpose: Mahuang decoction (MHD) is a classic famous traditional Chinese medicine and has various pharmacological effects, including anti-inflammation and anti-asthma. In this study, we aimed to investigate the potential protective effect of MHD against asthma and elucidated the underlying mechanism.
Materials and Methods: A mouse model of asthma was induced by ovalbumin (OVA) treatment, and then treated with MHD to evaluate its effect on the asthma. Gain- or loss-of-function approaches were performed in SP1 and FGFR3 to study their roles in asthma via measurement of airway inflammation, airway remodeling and airway smooth muscle cell (ASMC) proliferation-related factors.
Results: MHD reduced airway inflammation and remodeling. Additionally, MHD contributed to diminished expression of SP1, which was shown to repress airway inflammation and remodeling. Furthermore, SP1 bound to the FGFR3 promoter, resulting in the FGFR3 transcription promotion and ASMC proliferation. Conversely, FGFR3 knockdown abolished airway inflammation and remodeling, the mechanism of which was related to suppression of the PI3K/AKT signaling pathway. Meanwhile, MHD hindered airway inflammation and remodeling following asthma by suppressing the SP1/FGFR3/PI3K/AKT axis.
Conclusion: Taken together, MHD may retard airway inflammation and remodeling by suppressing the SP1/FGFR3/PI3K/AKT axis, which contributes to an extensive understanding of asthma and may provide novel therapeutic options for this disease.
Keywords: Mahuang decoction, asthma, SP1, FGFR3, PI3K/AKT signaling pathway, airway inflammation, airway remodeling
Introduction
Asthma is a highly heterogeneous disease, encompassing both atopic and non-atopic phenotypes,1 and frequently manifests with dyspnea, wheeze, chest tightness, and cough.2 Pediatric asthma, the most common chronic disease of childhood, causes a significant burden to the health care system.3 Airway inflammation and remodeling are among the most important pathological features of asthma.4 Therefore, identification of the specific molecular mechanism underlying airway inflammation and remodeling facilitates the development of treatment approaches for the management of asthma.
Mahuang decoction (MHD) is a traditional Chinese medicine composed of four different herbs: Ephedrae herba, Armeniacae semen, Cinnamomi ramulus and Glycyrrhizae radix, and has been widely used as a prescription for allergic reaction and inflammation for many years.5,6 MHD has been reported to be an effective treatment option for asthma due to its role in mitigating airway inflammation.7,8 A recent study has highlighted that modified MHD is capable of inhibiting inflammatory responses caused by cigarette smoke in human airway epithelial cells.6
SP1 exerts critical function in human diseases because of its modulation in genes associated with cellular processes in mammalian cells.9 A recent work revealed SP1 as an intensively associated hub gene in the integrated network of DNA methylation and gene expression influencing the asthma development.10 Additionally, the linkage of SP1 to the airway remodeling induced by WNT-5A has been documented.11 As previously confirmed, fibroblast growth factor receptor 3 (FGFR3) is a Sp-modulated gene in bladder cancer cells.12 Suppressed FGFR3 contributes to ameliorated airway inflammation and remodeling in an ovalbumin (OVA)-induced mouse model of chronic asthma.13 Meanwhile, PI3K/AKT is a well-established downstream regulatory kinase of FGFR3 which positively regulates its expression.14 Inhibition of the PI3K/AKT signaling pathway is critical for suppression of airway inflammation and remodeling of asthmatic mice.15 Therefore, we hypothesized that MHD might play a therapeutic role in preventing the development and progression of asthma via the SP1/FGFR3/PI3K/AKT axis. To address the hypothesis, we studied the effect of MHD on airway inflammation and remodeling during asthma and elucidated the underlying mechanism so as to provide guidance for the treatment of MHD on asthma. The experimental results suggested that MHD intervention confirmed a strong inhibitory action on asthma by suppressing the SP1/FGFR3/PI3K/AKT axis.
Materials and Methods
Ethics Statement
Animal experimentations were approved by the Ethics Committee of Changchun University of Traditional Chinese Medicine and conducted on the basis of the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Due efforts were created to limit animals’ pain.
Bioinformatics Analysis
GeneCards, CTD and DisGeNET databases were used to retrieve asthma-related genes, which were then intersected using the jvenn tool to identify the candidate targets. SymMsp database was adopted to retrieve the targets of MHD. The obtained targets of MHD were intersected with the asthma-related candidate targets to obtain the candidate targets of MHD in the treatment of asthma. The drug-target network was visualized employing Cytoscape 3.5.1 software. Functional enrichment analysis of the targets was then performed using Metascape database and the hTFtarget database was used to predict the downstream targets.
Asthma-related gene expression dataset GSE27876 was downloaded from GEO database. The dataset includes 5 normal samples and 5 asthma samples, equipped with the platform annotation file of GPL6480. The R language “limma” package was applied for differential analysis of gene expression with logFC > 1.5, p value < 0.05 as the threshold to screen significantly highly expressed genes.
Preparation of MHD
Ephedra sinica Stapf (approximately 9 g), Glycyrrhiza uralensis Fisch (approximately 3 g), Armeniacae Amarum Semen (approximately 1 g), and Cinnamomum cassia Presl (approximately 6 g) (the percentage of MHD depends on the clinical dose) were weighed and mixed. The mixture was then immersed in 8-fold volume of distilled water for 1 h, and decocted twice in boiling water for 330 min each time. The liquid was collected and centrifuged at 3000 g for 5 min and the supernatant was concentrated and dried in vacuum at 55°C. According to the original drying material, the yield of dried powder is 18.8%. The final dried powder of MHD was dissolved in distilled water to a final concentration of 2 g/mL. The samples were stored at 4°C (The storage time shall not exceed 2 months, and it shall be filtered and sterilized before storage). The dose of distilled water extract of MHD was expressed as the grams of original dry matter per kg body weight. The dose of distilled water extract of MHD (5, 10 and 20 g/kg) was given according to the body weight of animals.
High-Performance Liquid Chromatography (HPLC) Analysis of MHD Extract Components
Nine marker components of MHD (ephedrine HCl, amygdalin, 6-gingerol, glycyrrhizin, liquiritin apioside, liquiritin, cinnamaldehyde, cinnamic acid and coumarin) were analyzed by HPLC (Shimadzu, Corp., Kyoto, Japan). Chromatographic data were analyzed utilizing LabSolutions software (Version 5.54 SP3, Shimadzu). Next, the nine components were subjected to chromatographic separation using a SunFire C18 column. For simultaneous determination, 100 mg of the freeze-dried modified MHD powder dissolved in 20 mL of DW was extracted for 20 min at ambient temperature with the help of an ultra-sonicator (Branson 8510, Danbury, CT) and filtered through a 0.2-μm filter (PALL Life Sciences, MI).6
Establishment of OVA-Induced Asthma Mouse Models
Ninety 6–8 week-old healthy C57BL/6 wild-type (WT) female mice (weighing 20 ± 2 g; Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were housed in the animal experiment center of Shanghai Jiading Traditional Chinese Medicine Hospital with humidity of 40–60% and temperature of 21–27°C under a 12-h light/dark cycle (eat and drink freely). FGFR3 gene knockout (FGFR3−/−) mice were purchased from Cyagen Bioscience Inc. (Suzhou, Jiangsu, China). All mice were acclimatised for one week before experiment.
The WT mice were randomized into the control group (treated with saline, n = 10) and OVA group (treated with OVA, n = 80). Twenty FGFR3−/− mice were randomized into the Con + FGFR3−/− group (n = 10) and the OVA + FGFR3−/− group (n = 10). The mice in each group were numbered for noninvasive lung function test and preparation of lung tissue specimens and bronchoalveolar lavage fluid (BALF). OVA-treated mice were sensitized by intraperitoneal injection of 0.2 mL of sensitization solution containing 100 μg OVA (Sigma-Aldrich Chemical Company, St Louis, MO) and 2 mg aluminum hydroxide (Pierce Biotechnology Inc., Rockford, IL) on day 1 and day 14. Control mice were intraperitoneally injected with 0.2 mL of normal saline. On day 21, mice were anesthetized with 1% pentobarbital sodium followed by intranasal injection of 30 μg OVA.
As shown in Figure S1, 80 OVA-induced asthma model mice were randomized into (10 mice in each group): OVA group (intraperitoneal injection of OVA), OVA + MHD group (intraperitoneal injection of OVA and oral administration of 20 mg/g MHD), OVA + DEX group (peritoneal injection of OVA and oral administration of 3 mg/kg DEX [15032521, Anhui Jintaiyang Pharmaceutical Co., Ltd., Anhui, China]), OVA + sh-NC (peritoneal injection of OVA and intranasal instillation of adenovirus coated with negative control), OVA + sh-SP1 (peritoneal injection of ovalbumin and intranasal instillation of adenovirus coated with sh-SP1), OVA + MHD + oe-NC group (peritoneal injection of OVA, oral administration of 20 mg/g MHD and intranasal instillation of adenovirus coated with negative control), OVA + MHD + oe-SP1 group (peritoneal injection of OVA, oral administration of 20 mg/g MHD and intranasal instillation of adenovirus coated with oe-SP1), OVA + LY294002 group (peritoneal injection of OVA and intraperitoneal injection of 1.5 mg/kg LY294002 [Calbiochem, France Biochem, Meudon, France], a PI3K/AKT signaling pathway inhibitor).
From the 21st day, 30 min before the intranasal OVA treatment, the mice in the OVA + MHD group were given oral MHD treatment at a dose of 20 mg/g, the mice in the OVA + DEX group were intraperitoneally injected with 2 mg/kg DEX, and the mice in the OVA + LY294002 group were intraperitoneally injected with 1.5 mg/kg LY294002, all of which lasted 3 consecutive days until the 23rd day. The mice in the OVA + sh-NC, OVA + sh-SP1, OVA + MHD + oe-NC, OVA + MHD + oe-SP1 groups were instilled 30 min before intranasal OVA treatment at the 21st day by intranasal instillation of 10 μL 3.5×109 PFU adenovirus (Sangon Biotech, Shanghai, China). All adenoviruses were procured from Shanghai Sangon Bioengineering Co., Ltd. (Shanghai, China).
Measurement of Airway Hyperresponsiveness (AHR)
On the basis of AniRes 2005 animal pulmonary function system, within 24 h after the final OVA exposure, AHR of mice was tested by MeCh (Sigma) challenge. The respiratory rate was pre-set at 90/min, and the time ratio of expiration/inspiration was set as 20:10. AHR was then assessed using expiratory resistance (Re), inspiratory resistance (Ri), and the minimum value of Cdyn. R-areas of Ri and Re, the graph area between the peak value and baseline, and the trough of Cdyn were obtained for further analysis.
Preparation of Lung Tissue Specimens and BALF
Within 24 h after the final OVA exposure, the mice were anesthetized. BALF was implemented by the instillation of 0.8 mL cold PBS through the self-made trachea (Figure S2) into the lung and the liquid was withdrawn. This process was repeated for three times for collection of a total of 2 −5 mL liquid from each mouse. After that, the obtained BALF sample was centrifuged at 200 g and 4°C for 10 min and the supernatant was harvested and stored at −80°C for subsequent enzyme-linked immunosorbent assay (ELISA). The cell sediment was suspended in 200 μL of PBS, centrifuged, and subjected to Wright-Giemsa stain. The percentage of eosinophils in the BALF was calculated by counting 100 cells in randomly selected areas using an optical microscope (Olympus, Japan).
Hematoxylin-Eosin (HE) Staining
After BALF, mice were fixed for 5 min with 10% neutral formalin by inflation at 25 cm water pressure. Next, the left upper lobe lung tissue of mice was harvested and fixed in 4% paraformaldehyde for 24 h, paraffin-embedded and cut into 5-μm-thick sections. Ten bronchial cross-sections with diameters of 100–200 μm were randomly selected from each section under a light microscope (× 200). HE staining images were obtained using a microscope (Leica-DM2500, Germany) and analyzed using the Image Pro Plus 7.1 software (Media Cybernetics, Silver Spring, MD).
Periodic Acid-Schiff (PAS) Staining
This assay was conducted as previously reported.16 Paraffin sections were observed under a microscope to assess the pathological changes in lung tissues.
Masson’s Trichrome Staining
Paraffin sections were stained in hematoxylin and in ponceau-acid fuchsin solution, and hydrolyzed by 1% molybdophosphoric acid aqueous solution. Next, the sections were stained in 1% aniline blue or green light liquid for 5 min and treated with 1% glacial acetic acid aqueous solution for 5 s. Then, the sections were dehydrated in ascending series of alcohol and observed for the collagen deposition in the airway wall with the help of a microscope (blue: collagen fiber; red: muscle-fiber cytoplasm stained red; nuclei: blue-brown).
Immunohistochemistry (IHC)
Lung tissues of mice were paraffin-embedded and cut into 4-μm-thick sections, which were then submitted to antigen retrieval. Then, the sections were immunostained with primary antibodies anti-SP1 (ab227383, 1:100, Abcam Inc., Cambridge, UK) and anti-FGFR3 (MA5-32620, Thermo Fisher Scientific Inc., Waltham, MA) at 4°C overnight. The following day, the sections were incubated with biotinylated secondary antibody goat-anti rabbit IgG (1:1000, ab6721, Abcam) for 20 min, followed by additional incubation with HRP-streptavidin (Innova Biosciences) for 20 min. Following development with DAB, the sections were counterstained with hematoxylin and observed under a microscope (Leica-DM2500, Leica, Germany). Image-Pro Plus (version 7.1, Media Cybernetics) was adopted for quantitative analysis.
RT-qPCR
Total RNA was extracted from airway smooth muscle cell (ASMC) or mouse lung tissues using TRIzol reagents (Invitrogen, Carlsbad, California), with the quantity and concentration determined by ultraviolet visible light spectrometer (ND-1000, Nanodrop). The RNA was then reversely transcribed into cDNA employing the reverse transcription kit (RR047A, Takara Bio Inc., Otsu, Shiga, Japan). RT-qPCR was processed employing the SYBR® Premix Ex TaqTM II (Perfect Real Time) kit (DRR081, Takara) on an ABI 7500 instrument (Applied Biosystems, Foster City, CA). The primers are shown in Table S1. GAPDH was considered as a normalizer and the fold changes were calculated by the 2−ΔΔCt method.
Primary Mouse ASMC Isolation, Culture and Identification
The trachea and lung tissues of the mice were rinsed in D-Hanks/PBS solution with their outer membrane and connective tissues stripped off. The tissues were made into 3 mm sections which were washed by D-Hank’s liquid (HMK0011, Beijing Huamaike Biotechnology Inc., Beijing, China) and incubated with Dulbecco’s modified Eagle’s medium (DMEM) (SH30021, Beijing North TZ-Biotech Develop Inc., Beijing, China) supplemented with 0.15% collagenase in an electromagnetic stirrer (constant-temperature, 1008011909, Tianjin Shidanda Trade Inc., Tianjin, China) at 37°C for 30 min, followed by centrifugation at 100 g for 5 min. Next, the obtained precipitate mixed with the low-glucose DMEM appended to 20% fetal bovine serum (FBS) was adopted for cell suspension preparation. Then, the cell suspension was treated with 0.25% trypsin appended to 0.02% EDTA in a 37°C incubator, followed by reaction termination. Thereafter, the cells were centrifuged at 200 g for 5 min, resuspended in DMEM appended to 20% FBS, and cultured at 37°C in a 5% CO2 incubator. The solution was renewed once every two to three days. Under 80% confluence, cells were trypsinized and observed under a microscope. The cells were purified employing a differential adherence method with the cell purity of 100%. ASMCs at passage 5–8 were used for the subsequent experimentations. Finally, the primary ASMCs were identified with the help of immunocytochemistry staining.
Immunocytochemistry Staining
ASMCs were seeded onto the sterile cover glass to make 70% confluence, fixed in 4% paraformaldehyde for 20 min and immunolabeled with anti-α-SMA antibody (MA5-11544, Thermo Fisher Scientific) at 4°C overnight. The cells were then incubated with biotin-labeled secondary antibody rabbit anti-mouse IgG. The signal was detected by adding fast red, and then the cells were counterstained with modified Mayer’s hematoxylin. The cover glass was fixed with a crystal holder and visualized with Axio Vision software (Carl Zeiss, Inc, Thornwood, NY).
Cell Grouping and Transfection
ASMCs were isolated from control and OVA-treated mice. ASMCs from OVA-treated mice were then transfected with 50 ng/mL of plasmids (Sangon) of sh-NC, sh-SP1 and sh-FGFR3, or treated with 1% DMSO (Calbiochem) and 25 μM LY294002. ASMCs from control mice were used as the controls.
Immunofluorescence
Cell slides were washed with PBS, blocked with 10% BSA and probed at 4°C for 12–16 h with primary antibodies against SP1 (rabbit, 1:1000, ab227383, Abcam) and FGFR3 (rabbit, MA5-32620, Thermo Fisher Scientific). Then, the slide was re-probed with Alexa Fluor 647 donkey anti-rabbit IgG (Thermo Fisher Scientific) for 1 h at 25°C and then stained with 10 μg/mL 4’,6-diamidino-2-phenylindole (Sigma-Aldrich) for 15 min. Finally, the immunolabeled sections were observed under an Olympus BX51 fluorescence microscope (Olympus) or a laser scanning confocal fluorescence microscope (FV500, Olympus).
Chromatin Immunoprecipitation-Polymerase Chain Reaction (ChIP-PCR)
ChIP Kit (Thermo Fisher Scientific) was procured for this assay. Cells following treatments were collected and fixed with 1% formaldehyde to produce DNA-protein cross-linking. The cells were then submitted to ultrasonic treatment for making chromatin fragments. Next, cell lysate was incubated with the antibody against SP1 (rabbit, ab227383, 1:50, Abcam) to immunoprecipitate the complex. RT-qPCR was started to quantify ChIP products. The primer sequences of the FGFR3 promoter were as follows: forward: 5’-AGGCCCCATCAACAAAGGAG-3’ and reverse: 5’-GTGACCAACCCTCAGACCAGG-3’.
Dual-Luciferase Reporter Assay
The Jaspar database was applied to predict the binding site between SP1 and FGFR3 promoter sequences (about 1000 bp upstream of FGFR3 transcription initiation site). FGFR3 dual luciferase reporter gene vectors and mutant type (MUT) plasmids with SP1 binding sites were constructed respectively: pGL4.16-FGFR3-WT (−800/+1 bp), pGL4.16-FGFR3-MUT-1 (−800/-600 bp), pGL4.16-FGFR3-MUT-2 (−600/-400 bp), pGL4.16-FGFR3-MUT-3 (−400/-200 bp) and pGL4.16-FGFR3-MUT-4 (−200/+1 bp). The reporter plasmids were then co-transfected into HEK293T cells (cultured with MEM medium containing 10% FBS in a cell incubator with 5% CO2 at 37°C) with oe-SP1 and oe-NC plasmids. After 48 h, Dual-Luciferase® Reporter Assay System (E1910, Promega Corporation, Madison, WI) was adopted for quantifying the luciferase activity.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 kit (K1018, Apexbio) was procured for assessing cell proliferation. ASMCs in each group were seeded into 96-well plates with medium containing 100 μL 10% FBS at a density of 1×103 cells/well. After culture for 12, 24, 36 and 48 h, each well was added with 10 μL CCK-8 solution and cultured at 37°C for 2 h. Subsequently, the optical density values at 450 nm were determined by a microplate reader (51119080, Thermo Fisher Scientific). Each experiment was set up with 5 parallel wells and repeated three times independently.
Western Blot
The obtained total protein extracts were separated and electrotransferred onto polyvinylidene fluoride membranes which were then probed at 4°C overnight with the following diluted rabbit primary antibodies: SP1 (1:1000, ab227383, Abcam), FGFR3 (MA5-32620, Thermo Fisher Scientific), COL1A2 (1:2000, ab96723, Abcam), COL3A1 (1:3000, ab7778, Abcam), PI3K (1:1000, ab154598, Abcam), phosphorylated (p)-PI3K (1:1000, ab191606, Abcam), AKT (1:500, ab8805, Abcam), p-AKT (1:1000, ab38449, Abcam) and GAPDH (1:5000, ab194486, Abcam) as well as with the HRP-labeled secondary antibody (goat anti-rabbit, 1:5000, ab205718, Abcam) for 1.5 h at ambient temperature. The bands were visualized by addition of developing solution (NCI4106, Pierce, Rockford, IL) and band intensities were quantified using ImageJ 1.48 u software (Bio-Rad, Hercules, CA).
ELISA
By referring to the ELISA kit (Dakewe Biotech Company) instruction, the levels of the inflammatory factors interleukin (IL)-4, IL-6 and tumor necrosis factor-α (TNF-α) in the collected mouse BALF and the supernatant of ASMCs were detected accordingly.
Statistical Analysis
SPSS 21.0 statistical software (IBM Corp. Armonk, NY) was run for data analysis. Measurement data were described as mean ± standard deviation. Data between two groups were analyzed using unpaired t-test, while data among multiple groups were assessed using one-way analysis of variance (ANOVA), followed by Tukey’s post hoc tests. Repeated measures ANOVA was used for data comparison at different concentrations and two-way ANOVA for comparison of cell viability at different time points. p < 0.05 described statistical significance.
Results
MHD Inhibits Airway Inflammation and Remodeling of Asthmatic Mice
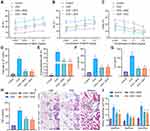
In order to verify the role of MHD in asthma, we first constructed a mouse asthma model induced by OVA. The mouse model was then treated with MHD and subjected to MeCh-stimulated AHR test. Ri and Re in the OVA-treated mice were found to be progressively increased over the MeCh dose, while the trough value of Cdyn was decreased. OVA exposure at each time point had a significant impact on Ri, Re and Cdyn, indicating the successful establishment of the mouse asthma model. In addition, MHD treatment markedly reduced the changes of lung function, with the effect similar to that of DEX treatment (Figure 1A-C).
The number of total cells and the percentage of eosinophils were increased in BALF of OVA-treated mice, while treatment with MHD or DEX caused an opposite result (Figure 1D and E). Moreover, ELISA data showed an increase of the levels of IL-4, IL-6 and TNF-α in the BALF of OVA-treated mice, while treatment with MHD or DEX decreased their levels (Figure 1F-H).
Analysis on the lung tissue using HE, PAS and Masson’s trichrome staining suggested severe peribronchial inflammatory infiltration, increased mucus secretion and collagen deposition and obviously thickened airway wall in lung tissues of OVA-treated mice, and conversely, treatment with MHD or DEX could reverse these results (Figure 1I and J).
These results suggested that MHD could inhibit airway inflammation of asthmatic mice.
SP1 is Highly Expressed in the Lung Tissues of Asthmatic Mice, and MHD Inhibits the Expression of SP1
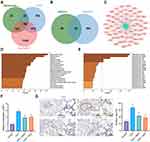
We then moved to explore the potential molecular targets of MHD in relieving airway inflammation and remodeling following asthma. GeneCards, CTD and DisGeNET databases were searched to retrieve the asthma-related genes, which were then intersected using the jvenn tool (Figure 2A). Following intersection analysis with the targets of MHD, 63 genes were obtained (Figure 2B). Cytoscape 3.5.1 was adopted to construct a regulatory network involving the drug-target (Figure 2C). Enrichment analysis using the Metascape database revealed that 63 genes may be related to reperfusion injury, inflammation, and asthma (Figure 2D), of which RELA, NFKB1, JUN, SP1 and STAT1 may be the key genes (Figure 2E).
The results of RT-qPCR and IHC demonstrated that in the lung tissues of OVA-treated mice, the expression of SP1 was increased but treatment with MHD or DEX led to opposite trend (Figure 2F and G).
Collectively, SP1 was highly expressed in the lung tissue of asthmatic mice, and MHD could inhibit the expression of SP1. Therefore, we chose SP1 as the target gene for subsequent experiments.
Silencing of SP1 Suppresses Airway Inflammation and Remodeling of Asthmatic Mice
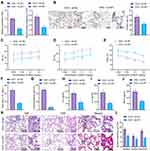
Next, we probed into the role of SP1 in airway inflammation and remodeling following asthma. Initial results identified reduced expression of SP1 in the lung tissues of OVA-treated mice treated with sh-SP1 (Figure 3A and B), indicating the efficiency of SP1 knockdown in mice. As shown in Figure 3C-G, AHR was abated and the number of total cells and the percentage of eosinophils were decreased in OVA-treated mice following SP1 knockdown.
Meanwhile, ELISA data showed declines in the levels of IL-4, IL-6 and TNF-α in the BALF of OVA-treated mice treated with sh-SP1 (Figure 3H-J). Further analysis on the lung tissues using HE, PAS and Masson’s trichrome stainings suggested that SP1 knockdown decreased peribronchial inflammatory infiltration, mucus secretion, collagen deposition and thickness of airway wall in lung tissues of OVA-treated mice (Figure 3K and L).
Cumulatively, silencing of SP1 could reduce airway inflammation and remodeling of asthmatic mice.
SP1 Promotes ASMC Cell Proliferation Through Transcriptional Activation of FGFR3
To elucidate the mechanism by which SP1 affects airway inflammation and remodeling during asthma, we first employed the hTFtarget database to predict the downstream target genes of SP1, which were then subjected to Venn diagram analysis with the significantly highly expressed genes obtained from the asthma-related dataset GSE27876. Seven genes were found at the intersection, including SEMA3F, ASB4, TOMM34, PTPRN, SEZ6, ERBB3, and FGFR3 (Figure 4A and B). RT-qPCR results further confirmed that FGFR3 showed the highest expression in the lung tissues of OVA-treated mice than the remaining genes (Figure 4C). It has been reported that inhibiting the expression of FGFR3 could reduce airway inflammation, and SP1 could bind to the FGFR3 promoter region.17,18 Therefore, we speculated that SP1 may promote airway inflammation and remodeling during asthma by modulating the transcription of FGFR3.
In addition, the expression of FGFR3 was detected to be elevated in the lung tissues of OVA-treated mice. However, SP1 knockdown inhibited the FGFR3 expression (Figure 4 and E). Jasper website predicted the binding site of SP1 to the FGFR3 promoter. Dual-luciferase reporter assay further showed that SP1 promoted the luciferase activity of FGFR3-WT, while inhibiting that of FGFR3-MUT-1, with no significant effects on the luciferase activity of FGFR3-MUT-2, FGFR3-MUT-3 and FGFR3-MUT-4 (Figure 4F), indicating that SP1 could target FGFR3 promoter Site1 and activate its transcription.
Immunocytochemistry staining analysis suggested the successful isolation of primary ASMCs from mouse lung tissues (Figure S3). ChIP-PCR results showed an increase in the enrichment of SP1 in the FGFR3 promoter region in AMSCs from OVA-treated mice (Figure 4G). Moreover, elevated expression of SP1, FGFR3, COL1A2 and COL3A1 was witnessed in AMSCs from OVA-treated mice, which was reversed following treatment with sh-SP1. Conversely, combined treatment with sh-SP1 and oe-FGFR3 resulted in upregulated FGFR3, COL1A2 and COL3A1 expression (Figure 4H and I). CCK-8 assay results demonstrated an upward trend in the proliferation of AMSCs from OVA-treated mice but SP1 knockdown impaired the proliferation. In the presence of both SP1 knockdown and FGFR3 overexpression, cell proliferation was noted to be increased (Figure 4J).
Therefore, SP1 could bind to the FGFR3 promoter and activate its transcription to stimulate ASMC proliferation and thus reduce airway inflammation and remodeling during asthma.
Down-Regulation of FGFR3 Expression Inhibits ASMC Proliferation and Airway Inflammation and Remodeling of Asthmatic Mice
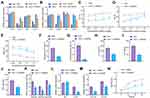
We then elucidated the role of FGFR3 transcription in airway inflammation and remodeling following asthma. AHR was abated and the number of total cells and the percentage of eosinophils were decreased in OVA-treated mice following FGFR3 knockdown (Figure 5A-E). In addition, the results of ELISA showed no difference in the levels of IL-4, IL-6 and TNF-α in the BALF between WT and FGFR3−/− mice after normal saline treatment. However, following OVA treatment, FGFR3−/− mice had decreased IL-4, IL-6 and TNF-α levels compared with WT mice (Figure 5F-H).
HE staining results showed no significant changes in the airway wall thickness of WT and FGFR3−/− mice after normal saline treatment; however, in the presence of OVA treatment, the airway wall thickness of FGFR3−/− mice was decreased in comparison with WT mice (Figure 5I). Next, we knocked down FGFR3 gene in primary ASMCs. We found reductions in the expression of FGFR3, COL1A2 and COL3A1 in AMSCs from OVA-treated mice treated with sh-FGFR3 (Figure 5J and K). CCK-8 assay results demonstrated a downward trend in the proliferation of AMSCs in the absence of FGFR3 (Figure 5L).
These results highlighted the ability of FGFR3 knockdown to reduce ASMC proliferation and the airway inflammation and remodeling.
Downregulation of FGFR3 Expression Inhibits ASMC Proliferation and Airway Inflammation and Remodeling of Asthmatic Mice by Inhibiting the PI3K/AKT Signaling Pathway
Previous literature has reported that FGFR3 activates the PI3K/AKT signaling pathway which is involved in airway inflammation and remodeling in asthmatic mice.19,20 We then proceeded to examine the mechanism of FGFR3 in the airway inflammation and remodeling via regulation of the PI3K/AKT signaling pathway. We identified downregulated phosphorylation levels of PI3K and AKT in the lung tissues of FGFR3−/− mice but no significant changes were observed in the total protein of PI3K and AKT (Figure 6A). In addition, the phosphorylation levels of PI3K and AKT were elevated in AMSCs from OVA-treated mice while FGFR3 knockdown resulted in opposite results (Figure 6B).
Treatment with LY294002 reduced AHR and the number of total cells and the percentage of eosinophils in OVA-treated mice (Figure 6C-G). Meanwhile, ELISA data showed lower levels of IL-4, IL-6 and TNF-α in the BALF of mice treated with OVA and LY294002 compared to mice treated with OVA alone (Figure 6H-J). Furthermore, the thickness of airway wall presented with a decline in lung tissues of OVA-treated mice in the presence of LY294002 (Figure 6K). In addition, in ASMCs, LY294002 diminished the AKT phosphorylation level (Figure 6L). The collagen deposition was noted to decreased in ASMCs of LY294002-treated OVA-sensitized mice (Figure 6M), and meanwhile, the proliferation of ASMCs was suppressed in response of LY294002 treatment (Figure 6N).
Thus, downregulation of FGFR3 could suppress the PI3K/AKT signaling pathway to repress ASMC proliferation and the resultant airway inflammation and remodeling.
MHD Inhibits Airway Inflammation and Remodeling of Asthmatic Mice by Inhibiting the SP1/FGFR3/PI3K/AKT Axis
Finally, we further validated the effect MHD on airway inflammation and remodeling during asthma through SP1-mediated FGFR3/PI3K/AKT axis. The results of Western blot analysis showed an enhancement in the PI3K and AKT phosphorylation levels in lung tissues of OVA-treated mice, while MTH treatment reversed the results, similar to that following DEX treatment (Figure 7A). Additionally, SP1 silencing led to reduced PI3K and AKT phosphorylation levels in lung tissues of OVA-treated mice (Figure 7B). These results suggested that either MHD or SP1 silencing suppressed activation of the PI3K/AKT signaling pathway in lung tissues of asthmatic mice.
Further data from RT-qPCR and IHC demonstrated upregulated SP1 and FGFR3 expression in the presence of both MHD and oe-SP1 (Figure 7C and D). Meanwhile, dual treatment with MHD and oe-SP1 enhanced AHR (Figure 7E-G), and increased the number of total cells and the percentage of eosinophils in OVA-treated mice (Figure 7H and I).
Meanwhile, ELISA results also showed augmented levels of IL-4, IL-6 and TNF-α in response to both MHD and SP1 overexpression (Figure 7J-L). Further analysis on the lung tissues using HE, PAS and Masson’s trichrome stainings suggested that combined treatment with MHD and oe-SP1 augmented peribronchial inflammatory infiltration, mucus secretion and collagen deposition in lung tissues of OVA-treated mice (Figure 7M). Moreover, Western blot analysis results revealed an elevation in the PI3K and AKT phosphorylation levels in lung tissues of OVA-treated mice co-treated with MHD and oe-SP1 (Figure 7N).
These lines of evidence suggested that MHD could inhibit airway inflammation and remodeling of asthmatic mice by downregulating SP1 expression and inhibiting the FGFR3/PI3K/AKT signaling pathway.
Discussion
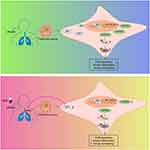
Asthma represents a chronic airway inflammatory disease mainly associated with heterogeneity.21 MHD, a well-known traditional Chinese medicine prescription, has been extensively applied for relieving the cold, influenza, cough, acute bronchitis, asthma and other pulmonary diseases.22,23 Here, this study suggested that MHD could potentially decrease SP1 expression and disrupted FGFR3-dependent PI3K/AKT signaling pathway activation, consequently preventing airway inflammation and remodeling, therefore delaying asthma progression (Figure 8).
It was noted in our work that MHD could alleviate airway inflammation and remodeling in mice with asthma. Shegan-MHD is capable of reducing AHR and attenuating the pulmonary infiltration of CD3 and CD4 T cells.24 MHD can also inhibit the pulmonary inflammation, evidenced by a marked inhibition of IL-1β, IL-6, and TNF-α in the BALF of cigarette smoke- and lipopolysaccharide-exposed mouse models through suppression of Erk phosphorylation.6 Additionally, the ameliorative role of MHD in bronchial asthma symptoms has also been demonstrated,25 which is partly in line with our findings. What’s more, MHD inhibits the release of inflammatory cytokines and thus inhibits the occurrence of asthma by down-regulating the phosphorylation level of STAT1.26 Airway inflammation is a main pathological feature of asthma and current therapeutic interventions focus primarily on resolving inflammation.27 Airway remodeling can be defined as the changes in the type, quantity and nature of airway wall compositions and their organization, which is also considered a most important pathological feature of asthma.28 Airway remodeling is a consequence of repeated inflammatory injury and repair of the respiratory tract.7 Meanwhile, many traditional Chinese medicines, including shegan MHD, have a regulatory role in the airway remodeling (https://pesquisa.bvsalud.org/portal/resource/pt/wpr-468274). Ephedra, one of the components of MHD, is an effective treatment for asthma due to its multi-target and multi-pathway functions.29 These findings demonstrate the protective effect of MHD against asthma by mitigating airway inflammation and remodeling.
The present study further identified the downregulated expression of SP1 was involved in the therapeutic effect of MHD on the asthma. A majority of traditional Chinese medicines can act as Sp1 antagonists to suppress its expression, such as Licochalcone A, Xiaoji decoction and Guifu Dihuang pill.30–32 This study represents the first report to reveal the inhibiting role of MHD in the expression of SP1. In line with our results, SP1 expression was previously detected to be abundant in primary bronchial epithelial cells from subjects with severe asthma and potentiated the airway remodeling associated with H1N1 infection.33 In addition, siRNA-mediated silencing of SP1 can prevent MMP1 expression,34 the activation of which contributes to ASMC proliferation and the subsequent asthma severity,35 suggesting the inhibiting effect of SP1 knockdown on the asthma progression. Due to the lack of available literature, the regulation of MHD in the SP1 expression requires further investigation.
Furthermore, our study unfolded that SP1 bound to the FGFR3 promoter and induced a resultant increase of transcriptional activation of the FGFR3 promoter. Consistently, SP1 has been identified to bind to the FGFR3 promoter and result in a significant increase in the transcriptional activity of the FGFR3 promoter.36 Additionally, transfection of a small inhibitory RNA cocktail (iSp) containing small inhibitory RNAs targeted to Sp1 (iSp1), Sp3 (iSp3) and Sp4 (iSp4) has been proved to decrease FGFR3 protein levels.12 Inhibited FGFR3 expression aids in the lowered AHR and expression of factors engaged in airway inflammation and remodeling in a mouse model of asthma.17 These findings offered evidence validating the inhibiting effect of SP1 knockdown on the airway inflammation and remodeling by inhibiting the transcriptional activity of the FGFR3 promoter. Furthermore, siRNA-mediated silencing of FGFR3 diminishes the expression of PI3K and AKT phosphorylation level in the context of subarachnoid hemorrhage.37 Meanwhile, suppression of the PI3K/AKT signaling pathway due to miR-221 inhibition in a murine asthma model has been shown to reduce AHR, mucus metaplasia, airway inflammation and remodeling.38 An increase of ASMCs is a hallmark of airway remodeling in asthma, and notably, inhibition of the PI3K/AKT signaling pathway has demonstrated an inhibitory role in the ASMC proliferation.39 Therefore, it might be plausible to suggest that FGFR3 knockdown reduced ASMC proliferation and airway inflammation and remodeling via PI3K/AKT signaling pathway inactivation.
Conclusion
Collectively, the key findings of this study provided evidence validating that MHD could downregulate the expression of SP1 and inhibit transcription of FGFR3, thereby resulting in inactivation of the PI3K/AKT signaling pathway. By this mechanism, MHD inhibited ASMC proliferation, resulting in the alleviation of the resultant airway inflammation and remodeling in mice with asthma. These findings emphasize a novel aspect of the role of MHD in asthma and inspire us to develop a novel strategy based on this drug reagent. However, further studies are necessitated to focus on clinical samples collected from patients with asthma for further validation of the therapeutic effects of MHD against OVA-induced asthma.
Data Sharing Statement
The datasets analysed during the current study are available.
Funding
This study was supported by Bronchial asthma in children: -Construction of dominant diseases (ZY (2018-2020)-ZYBZ-40); National Natural Science Foundation of China (NO: 81974579).
Disclosure
No competing interest in our study.
References
1. Sobeih AA, Behairy OG, Abd Almonaem ER, Mohammad OI, Mn Abdelrahman A. Clusterin in atopic and non-atopic childhood asthma. Scand J Clin Lab Invest. 2019;79(6):368–371. doi:10.1080/00365513.2019.1624976
2. Tyler SR, Bunyavanich S. Leveraging -omics for asthma endotyping. J Allergy Clin Immunol. 2019;144(1):13–23. doi:10.1016/j.jaci.2019.05.015
3. Simoneau T, Cloutier MM. Controversies in Pediatric Asthma. Pediatr Ann. 2019;48(3):e128–e134. doi:10.3928/19382359-20190226-01
4. Fehrenbach H, Wagner C, Wegmann M. Airway remodeling in asthma: what really matters. Cell Tissue Res. 2017;367(3):551–569. doi:10.1007/s00441-016-2566-8
5. Zhao Y, Jia L, Wang J, Zou W, Yang H, Xiao X. Cold/hot pad differentiating assay of property differences of Mahuang and Maxingshigan decoctions. Pharm Biol. 2016;54(7):1298–1302. doi:10.3109/13880209.2015.1057650
6. Ko JW, Seo CS, Shin NR, et al. Modificated Mahuang-Tang, a traditional herbal medicine suppresses inflammatory responses induced by cigarette smoke in human airway epithelial cell and mice. Phytomedicine. 2019;59:152777. doi:10.1016/j.phymed.2018.11.037
7. He Y, Lou X, Jin Z, Yu L, Deng L, Wan H. Mahuang decoction mitigates airway inflammation and regulates IL-21/STAT3 signaling pathway in rat asthma model. J Ethnopharmacol. 2018;224:373–380. doi:10.1016/j.jep.2018.06.011
8. Huang P, Tang Y, Li C, et al. Correlation study between the pharmacokinetics of seven main active ingredients of Mahuang decoction and its pharmacodynamics in asthmatic rats. J Pharm Biomed Anal. 2020;183:113144. doi:10.1016/j.jpba.2020.113144
9. O’Connor L, Gilmour J, Bonifer C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J Biol Med. 2016;89(4):513–525.
10. Kim JS, Shin IS, Shin NR, Nam JY, Kim C. Genomewide analysis of DNA methylation and gene expression changes in an ovalbumininduced asthma mouse model. Mol Med Rep. 2020;22(3):1709–1716. doi:10.3892/mmr.2020.11245
11. Kumawat K, Menzen MH, Slegtenhorst RM, Halayko AJ, Schmidt M, Gosens R. TGF-beta-activated kinase 1 (TAK1) signaling regulates TGF-beta-induced WNT-5A expression in airway smooth muscle cells via Sp1 and beta-catenin. PLoS One. 2014;9(4):e94801. doi:10.1371/journal.pone.0094801
12. Chadalapaka G, Jutooru I, Safe S. Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis. 2012;33(4):886–894. doi:10.1093/carcin/bgs102
13. Lee HY, Hur J, Kim IK, et al. Effect of nintedanib on airway inflammation and remodeling in a murine chronic asthma model. Exp Lung Res. 2017;43(4–5):187–196. doi:10.1080/01902148.2017.1339141
14. Starska K, Forma E, Lewy-Trenda I, Stasikowska-Kanicka O, Skora M, Brys M. Fibroblast growth factor receptor 1 and 3 expression is associated with regulatory PI3K/AKT kinase activity, as well as invasion and prognosis, in human laryngeal cancer. Cell Oncol. 2018;41(3):253–268. doi:10.1007/s13402-017-0367-z
15. Li X, Zhou L, Zhang Z, Liu Y, Liu J, Zhang C. IL-27 alleviates airway remodeling in a mouse model of asthma via PI3K/Akt pathway. Exp Lung Res. 2020;46(3–4):98–108. doi:10.1080/01902148.2020.1740356
16. Liu Y, Zhang H, Ni R, Jia WQ, Wang YY. IL-4R suppresses airway inflammation in bronchial asthma by inhibiting the IL-4/STAT6 pathway. Pulm Pharmacol Ther. 2017;43:32–38. doi:10.1016/j.pupt.2017.01.006
17. Lee J, Rhee CK, Lee JH, et al. Effect of nintedanib on airway inflammation in a mouse model of acute asthma. J Asthma. 2020;57(1):11–20. doi:10.1080/02770903.2018.1544641
18. Perez-Castro AV, Wilson J, Altherr MR. Genomic organization of the mouse fibroblast growth factor receptor 3 (Fgfr3) gene. Genomics. 1995;30(2):157–162. doi:10.1006/geno.1995.9890
19. Ahn SY, Chang YS, Sung DK, Sung SI, Park WS. Hypothermia broadens the therapeutic time window of mesenchymal stem cell transplantation for severe neonatal hypoxic ischemic encephalopathy. Sci Rep. 2018;8(1):7665. doi:10.1038/s41598-018-25902-x
20. Campa CC, Silva RL, Margaria JP, et al. Inhalation of the prodrug PI3K inhibitor CL27c improves lung function in asthma and fibrosis. Nat Commun. 2018;9(1):5232. doi:10.1038/s41467-018-07698-6
21. Perlikos F, Hillas G, Loukides S. Phenotyping and Endotyping Asthma Based on Biomarkers. Curr Top Med Chem. 2016;16(14):1582–1586. doi:10.2174/1568026616666150930120803
22. Chen L, Yao C, Li J, et al. Systematic characterization of chemical constituents in Mahuang decoction by UHPLC tandem linear ion trap-Orbitrap mass spectrometry coupled with feature-based molecular networking. J Sep Sci. 2021;44(14):2717–2727. doi:10.1002/jssc.202100121
23. He Y, Zhu Y, Zhang R, Ge L, Wan H. Simultaneous quantification of nine major active components in traditional Chinese prescription Mahuang decoction and the influence of herbal compatibility on their contents. Pharmacogn Mag. 2014;10(Suppl 1):S72–79. doi:10.4103/0973-1296.127346
24. Lin CC, Wang YY, Chen SM, et al. Shegan-Mahuang Decoction ameliorates asthmatic airway hyperresponsiveness by downregulating Th2/Th17 cells but upregulating CD4+FoxP3+ Tregs. J Ethnopharmacol. 2020;253:112656. doi:10.1016/j.jep.2020.112656
25. Jiao J, Wu J, Wang J, et al. Ma Huang Tang ameliorates bronchial asthma symptoms through the TLR9 pathway. Pharm Biol. 2018;56(1):580–593. doi:10.1080/13880209.2018.1517184
26. Lim HS, Seo CS, Jin SE, et al. Ma Huang Tang Suppresses the Production and Expression of Inflammatory Chemokines via Downregulating STAT1 Phosphorylation in HaCaT Keratinocytes. Evid Based Complement Alternat Med. 2016;2016:7831291. doi:10.1155/2016/7831291
27. Banno A, Reddy AT, Lakshmi SP, Reddy RC. Bidirectional interaction of airway epithelial remodeling and inflammation in asthma. Clin Sci (Lond). 2020;134(9):1063–1079. doi:10.1042/CS20191309
28. Boulet LP. Airway remodeling in asthma: update on mechanisms and therapeutic approaches. Curr Opin Pulm Med. 2018;24(1):56–62. doi:10.1097/MCP.0000000000000441
29. Huang XF, Cheng WB, Jiang Y, et al. A network pharmacology-based strategy for predicting anti-inflammatory targets of ephedra in treating asthma. Int Immunopharmacol. 2020;83:106423. doi:10.1016/j.intimp.2020.106423
30. Bai H, Zhou M, Zhou H, et al. Licochalcone A suppresses Sp1 expression with potential anti-myeloma activity. Cancer Commun. 2021;41(11):1239–1242. doi:10.1002/cac2.12223
31. Zhao S, Wu J, Tang Q, et al. Chinese herbal medicine Xiaoji decoction inhibited growth of lung cancer cells through AMPKalpha-mediated inhibition of Sp1 and DNA methyltransferase 1. J Ethnopharmacol. 2016;181:172–181. doi:10.1016/j.jep.2016.01.041
32. Zhang H, Yu W, Ji L, et al. Guifu Dihuang Pills Ameliorated Mucus Hypersecretion by Suppressing Muc5ac Expression and Inactivating the ERK-SP1 Pathway in Lipopolysaccharide/Cigarette Smoke-Induced Mice. Evid Based Complement Alternat Med. 2021;2021:9539218. doi:10.1155/2021/9539218
33. Moheimani F, Koops J, Williams T, et al. Influenza A virus infection dysregulates the expression of microRNA-22 and its targets; CD147 and HDAC4, in epithelium of asthmatics. Respir Res. 2018;19(1):145. doi:10.1186/s12931-018-0851-7
34. Lee KE, Jee HM, Hong JY, et al. German Cockroach Extract Induces Matrix Metalloproteinase-1 Expression, Leading to Tight Junction Disruption in Human Airway Epithelial Cells. Yonsei Med J. 2018;59(10):1222–1231. doi:10.3349/ymj.2018.59.10.1222
35. Naveed SU, Clements D, Jackson DJ, et al. Matrix Metalloproteinase-1 Activation Contributes to Airway Smooth Muscle Growth and Asthma Severity. Am J Respir Crit Care Med. 2017;195(8):1000–1009. doi:10.1164/rccm.201604-0822OC
36. McEwen DG, Ornitz DM. Regulation of the fibroblast growth factor receptor 3 promoter and intron I enhancer by Sp1 family transcription factors. J Biol Chem. 1998;273(9):5349–5357. doi:10.1074/jbc.273.9.5349
37. Okada T, Enkhjargal B, Travis ZD, et al. FGF-2 Attenuates Neuronal Apoptosis via FGFR3/PI3k/Akt Signaling Pathway After Subarachnoid Hemorrhage. Mol Neurobiol. 2019;56(12):8203–8219. doi:10.1007/s12035-019-01668-9
38. Pan J, Yang Q, Zhou Y, et al. MicroRNA-221 Modulates Airway Remodeling via the PI3K/AKT Pathway in OVA-Induced Chronic Murine Asthma. Front Cell Dev Biol. 2020;8:495. doi:10.3389/fcell.2020.00495
39. Liu Y, Li X, He C, et al. Emodin ameliorates ovalbumin-induced airway remodeling in mice by suppressing airway smooth muscle cells proliferation. Int Immunopharmacol. 2020;88:106855. doi:10.1016/j.intimp.2020.106855
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.