Back to Journals » International Journal of General Medicine » Volume 16
MAGED4B is a Poor Prognostic Marker of Stomach Adenocarcinoma and a Potential Therapeutic Target for Stomach Adenocarcinoma Tumorigenesis
Authors Luo WZ, Li X, Wu XX, Shang YW, Meng DH, Chen YL , Zhang QS
Received 21 December 2022
Accepted for publication 26 April 2023
Published 6 May 2023 Volume 2023:16 Pages 1681—1693
DOI https://doi.org/10.2147/IJGM.S401507
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Wen-Zhao Luo,1,2 Xian Li,2 Xiu-Xia Wu,2 Yi-Wan Shang,1,3 Dan-Hua Meng,1,3 Yu-Long Chen,1,3 Qin-Sheng Zhang2
1School of Basic Medicine (Zhongjing School), Henan University of Chinese Medicine, Zhengzhou, Henan Province, 45000, People’s Republic of China; 2Department of Hepatobiliary and Spleen Stomach, Henan Province Hospital of Traditional Chinese Medicine, Zhengzhou, Henan Province, 450000, People’s Republic of China; 3Academy of Chinese Medical Sciences, Henan University of Chinese Medicine, Zhengzhou, Henan Province, 45000, People’s Republic of China
Correspondence: Yu-Long Chen, School of Basic Medicine (Zhongjing School), Henan University of Chinese Medicine, No. 156 East Jinshui Road, Jinshui District, Zhengzhou, Henan Province, 45000, People’s Republic of China, Email [email protected] Qin-Sheng Zhang, Henan Province Hospital of Traditional Chinese Medicine, Department of Hepatobiliary and Spleen Stomach, No. 6 Dongfeng Road, Jinshui District, Zhengzhou, Henan Province, 450000, People’s Republic of China, Email [email protected]
Background: Gastric cancer is the second most common cause of cancer death worldwide with poor overall prognosis. It is important to study the molecular mechanism of stomach adenocarcinoma (STAD). MAGED4B, a member of the melanoma antigen gene (MAGE) family, is highly expressed in many tumor cells and is associated with tumor progression. Its prognostic value in and the function of the encoded protein are still unclear.
Methods: The data of 415 STAD tissues was retrieved from TCGA database, and the expression level of MAGED4B mRNA was evaluated. The correlation between the expression of MAGED4B mRNA and the progression free survival (PFS) time of STAD patients was evaluated by Kaplan Meier analysis. The STAD cell lines with overexpressed and silent MAGED4B were constructed, and the effects of MAGED4B on the viability, migration and proliferation were evaluated by the CCK-8, scratch test and EDU test. The flow cytometry was used to detect apoptosis with overexpressed and silent MAGED4B under the cisplatin treatment, and WB was used to detect the expressions of related proteins, such as TNF-α.
Results: The expression level of MAGED4B mRNA in the STAD tissues was higher than that in the normal tissues, and its high expression was related to poor PFS. The overexpression of MAGED4B in the STAD cell lines can promote the vitality, motility and proliferation of the STAD cells, while the silencing of MAGED4B can inhibit the above three cell functions of the STAD cells. The overexpression of MAGED4B can reduce the cisplatin induced apoptosis and increase the cisplatin IC50; the silencing of MAGED4B can promote the cisplatin induced apoptosis and reduce the cisplatin IC50. The overexpression of MAGED4B reduced the protein levels of TRIM27 and TNF- α.
Conclusion: MAGED4B could be a valuable prognostic biomarker and a therapeutic target for gastric adenocarcinoma of great interest.
Keywords: stomach adenocarcinoma, MAGED4B, prognosis, biomarkers
Introduction
Gastric cancer (GC) is the fifth most common cancer and the second leading cause of cancer death in the world. It is a complex disease caused by the interaction of the environment and related factors of the host. The main reasons for its high mortality include its silent nature, terminal clinical manifestations, and potential biological and genetic heterogeneity.1–3 Stomach adenocarcinoma (STAD) is a typical example of heterogeneous malignant diseases, which has different subtypes and clinical behaviors, and is affected by a variety of genetic and environmental factors. The accurate diagnosis and individualized treatment of STAD are serious challenges in modern medicine.2 The factor closely related to the prognosis of STAD is the tumor staging.4,5 In addition, the tumor location, tumor size, histological type, grading, depth of tumor invasion, regional lymph node involvement, distal metastasis and other factors are also reported to be related to the prognosis of STAD.6–8 In the last 10 years, the genetic changes of many genes have been found in gastric cancer. These genes play an important role in many cellular functions, such as signal transduction, proliferation and invasion.
The melanoma antigen gene (MAGE) proteins are a class of cancer testis antigens (CTAs), which are classified into type I and type II genes according to different gene structures and tissue specific gene expressions.9 Type I is expressed in the germ line cells and tumors, but not in the normal cells. Type II is widely expressed in various tissues.10–12 The MAGE proteins have been proved to be a good target for cancer immunotherapy. MAGED4B (type II) has overexpression in more than 50% of oral squamous cell carcinoma (OSCC) tissues, and the expression of MAGED4B is related to lymph node metastasis and poor survival.13 The OSCC cell lines with overexpressed MAGED4B can promote cell migration in vitro and present increased cell growth both in vitro and in vivo, which also have stronger resistance to apoptosis. The overexpression of MAGED4B is a driving factor for the occurrence of oral cancer, which can be used as a potential therapeutic target of OSCC.13 In addition, studies have shown that MAGED4B, as CTAs in clinical practice, is closely related to the poor prognosis of cancer patients, showing its potential as a diagnostic biomarker and a target for the discovery of new anti-cancer drugs.14
In this study, we analyzed the STAD expression profile data from The Cancer Genome Atlas (TCGA), and found that the expression level of MAGED4B mRNA was higher in the STAD tissues than that in the normal tissues, and its high expression was associated with the poor progression free survival (PFS) time. The gene overexpression/silencing experiments showed that MAGED4B may play a key role in the tumorigenesis of stomach adenocarcinoma cells by regulating the activity, movement and proliferation of stomach adenocarcinoma cells. In conclusion, MAGED4B could be a valuable prognostic biomarker and a cancer therapy target of great interest for stomach adenocarcinoma.
Materials and Methods
TCGA Data Analysis
The TCGA-STAD and corresponding clinical data of 415 STAD samples and 34 normal samples were all downloaded to The Cancer Genome Atlas (TCGA) library (https://gdc-portal.nci.nih.gov/).
Specimen Collection From stomach Adenocarcinoma Patients Receiving New Adjuvant Therapy
In this study, 76 cases of STAD samples and their matched normal tissues came from the Henan Province Hospital of Traditional Chinese Medicine (Mar, 2019 to Sep 2021). All patients had not received any chemotherapy, radiotherapy or immunotherapy before surgery. All research complied with The Declaration of Helsinki. This study was approved by the Ethics Committee of the Henan Province Hospital of Traditional Chinese Medicine, and all patients signed a written informed consent before surgery.
Immunohistochemistry Analysis
The SignalStay Boost IHC test kit was used to dye IHC according to the manufacturer’s instructions.15 To put it simply, the STAD chips were dewaxed in xylene, dehydrated by gradient ethanol and then rehydrated. Next, the Target Retrieval Solution (Dako, CA, USA) was used to extract antigen, and the 10% goat serum was used to block non-specific binding. The sections were incubated with the primary antibody anti-MAGED4B (TA344751, OriGene Technologies, Inc., USA) at 4 ℃ overnight, and then with the secondary antibody Goat anti-Rabbit IgG (31460, ThermoFisher, USA) to incubate for 1 hour. In the next step, the SignalStay DAB Substrate Kit (CST, USA) was used to stain nucleus.
Cell Lines and Cell Culture
All Gastric cell lines (AGS, BGC, HGC-27, SGC-7901, MGC-803) were purchased from the Nanjing Cobioer Biosciences CO.LTD (Nanjing, China, AGS, CBP60476; BGC, CBP60832; HGC-27, CBP60480; SGC-7901, CBP60500; MGC-803, CBP60485). All these cells were cultured in RPMI-1640 with 10% fetal bovine serum, and placed in the incubator (37°C, 5% CO2).
Construction of Cell Lines with Overexpressed/Silent MAGED4B
According to the manufacturer’s instructions, the MAGED4B Human shRNA Lentiviral Particle (TL303375V, OriGene Technologies, Inc., USA) was used to knock out the MAGED4B genes from the HGC-27 cells. The untreated cells (NC) were used as negative controls, and the cells without gene knockout received the same treatment and used as the internal references. The overexpression of MAGED4B in the BGC cells was achieved through the transfection of the MAGED4B overexpression plasmids with the final concentration of 1 µg/mL constructed with GenePharma, and the transfection was performed using the Lipofectamine® 2000 reagent (Invitrogen, Waltham, USA).
Fluorescence Quantitative PCR
According to the manufacturer’s instructions, the total RNA was extracted from tissues and treated cells (overexpressed/silent MAGED4B cell lines and their control cells) using the TRIzol reagent (Invitrogen, USA) and the purity and concentration of total RNA were determined by ultraviolet spectrophotometry. Then, the SuperScript III (Invitrogen) was used to synthesize cDNA according to the manufacturer’s instructions. The primer sequences of MAGED4B are: qF: 5 - GCAACGACTATGAAGAAT-3, qR: 5 - CTCTGAGATATGGATGT-3. The QRT PCR was performed in the ABI PRISM 7000 fluorescence quantitative PCR system (Applied Biosystems, Foster City, CA, USA) using the standard SYBR Green PCR kit (Takara, Dalian, China). QRT-PCR procedure: denaturing (95 °C for 5 seconds), annealing (55 °C for 30 seconds) and extending (72 °C for 30 seconds), it was repeated for 40 cycles. The 2−ΔΔCt method was adopted to analyze the relative expression of genes.
Scratch Test
The single-layer wound healing experiment was conducted according to the method in.16 In short, the cell suspension was inoculated into a 6-well plate, 1 mL in each well, the cell density was approximately 1×105, and it was cultured for 24 h. After cell adhesion, the RPMI-1640 culture medium was added to form monolayer cells; the 10 μL sterile tip was used to vertically scratch on the monolayer cells; then, PBS was added slowly along the wall to wash the exfoliated cells; next, the specimen was placed in an incubator with 5% CO2 for incubation under 37°C, and it was observed and photographed at 0 h and 24 h.
CCK-8 Assay
The Cell Counting Kit-8 (# CA1210, Beijing Solarbio Science &T echnology Co., Ltd) was used to detect the cell viability according to the instructions. In short, the stomach adenocarcinoma cells with overexpressed/silent MAGED4B and their control cells in logarithmic growth phase were inoculated into the 96-well plate, and the cell density was about 4×104 cells/well, 100 μL in each well, and the cells were incubated in the 5% CO2 incubator at 37 °C for 24 hours. Then, 10 μL CCK-8 solution was added to the cells, and air bubbles were avoided. Next, the cells added with the CCK-8 solution were cultured for 3–4 hours, and then the absorbance at 450 nm was measured with the microplate reader.
The silent or overexpressed cells were inoculated into the 96-well plate. The cisplatin of different concentrations (32, 16, 8, 4, 2, and 1 μg/mL) was used to treat the cells for 24 h, and then the cell viability of each well was determined according to the above CCK8 method. The regression model in SPSS 17.0 (SPSS Inc., Chicago, USA) was used for calculation of IC50.
EDU Experiment
The Cell Light EdU Apollo 567 In Vitro Kit (# C10310, Guangzhou RiboBio Co., Ltd.) was used to detect the cell proliferation according to the instructions. In short, the cells in logarithmic growth period were inoculated into the 96-well plate with 4×103 −1×105 cells per well, and then cultured to the normal growth stage. Then, the EdU solution was diluted using the complete cell medium at the ratio of 1000:1 to prepare the 50 μ M EdU medium; 100 μL of 50 μM EdU medium was added to each well to incubate for 2h, and then, the medium was discarded; PBS was used to wash the cells for 1–2 times. Then, 50 μL cell fixation solution was added to each well, and the cells were incubated at room temperature for 30 min; the fixation solution was discarded, 50 μL glycine (2 mg/mL) was added dropwise to each well, and the cells were incubated on the decolorization shaker for 5 min; the waste liquid was discarded, 100 μL PBS was used to wash the cells on the decolorization shaker for 5 min, and the waste liquid was discarded; next, 100 μL penetrant containing 0.5% TritonX-100 was added to each well, and the cells were incubated on the decolorization shaker for 10 min. Then, the cells were washed using PBS for 5 min; 100 μ L 1 × Apollo dye solution, was added, the cells were incubated at room temperature and away from light for 30 min on the decolorization shaker, and the waste liquid was discarded; 100 μL penetrant was added to the decolorization shaker dropwise to incubate for 10 min, the waste liquid was discarded, and this procedure was repeated twice; next, 100 μL methanol was added to clean the slide for 5 min, and then PBS was used to wash for 5 min; 100 μL DAPI dye reaction solution was added to the slide dropwise, it was incubated on the decolorization shaker at room temperature and away from light for 30 min, the waste liquid was discarded, and it was washed with PBS for three times; after using the Fluoromount for mounting, five 200× fields of view were randomly chosen under the fluorescence microscope to observe and take photos; the Image Pro Plus 6.0 software was used to process the data.
WB
The cell lysates were extracted using the RIPA (Beyotime, Beijing) lysis buffer. The protein concentration was measured using the standard bovine serum albumin (BSA) protein kit (Thermo, USA). The samples were separated by SDS-PAGE, transferred to the PVDF membrane (Millipore, USA), and sealed in the 5% skimmed milk for 1h at room temperature. The PVDF membrane was incubated with the first antibody (MAGED4B: 1:1000, TA344751; TRIM27: 1:2000, CF803910; TNF-α: 1:500, AB0109-100, OriGene Technologies, Inc., USA) at 4 °C overnight, and then incubated with secondary antibodies (31460, 31430, 31402, ThermoFisher, USA) at 4 ℃ for 1h. Then, it was displayed with the ECL chemiluminescence detection system. The protein bands were quantified by density measurement and analysis using the Quantity One software (Bio Rad, Hercules, CA, USA), and the data was analyzed using the GraphPad Prism software (La Jolla, CA, USA).
Data Analysis
The statistical analysis was conducted using GraphPad Prism 9 (San Diego, CA, USA). Comparison among groups was performed using the one-way ANOVA analysis, the correlation between the MAGED4B expression and the clinical characteristics was evaluated via the χ2 test, the effect of MAGED4B expression level on progression free survival (PFS) was analyzed online through UALCAN (https://ualcan.path.uab.edu/ index.html) and the Kaplan Meier and log rank tests were used for survival analysis. Cox regression analyses were conducted to estimate independent prognostic factors for PFS. If P < 0.05, it was considered as statistically significant.
Results
MAGED4B is Up-Regulated in Gastric Adenocarcinoma
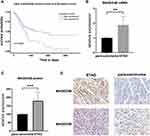
In the TCGA-STAD database, the expression of MAGED4B in the STAD samples (N=415) was not significantly different from that in the non-tumor samples (N=34). However, among the STAD patients, the patients with high expression of MAGED4B showed poor progression free survival (PFS) (Figure 1A). The RT qPCR was used to detect the transcript expression level of MAGED4B in 76 STAD clinical samples. The results showed that the expression of MAGED4B was significantly up-regulated compared with the para-carcinoma tissues (P<0.01) (Figure 1B). Then, the protein expression level of MAGED4B in the STAD clinical samples was detected by IHC, and the expression of MAGED4B in the stomach adenocarcinoma samples was significantly up-regulated compared with that in the para-carcinoma tissues (P<0.001) (Figure 1C and D). Next, the relationship between the expression of MAGED4B and the main clinicopathologic characteristics of stomach adenocarcinoma patients was analyzed, so as to evaluate the impact of MAGED4B on the progression of stomach adenocarcinoma. The results are shown in Table 1. The expression of MAGED4B was not related to gender, but related to age and the histologic type of tumor.
 |
Table 1 Relationship Between MAGED4B Expression and Clinical Characteristics in Stomach Adenocarcinoma |
Next, we selected five different STAD cell lines (AGS, BGC, HGC-27, SGC-7901, MGC-803) to detect the protein expression of MAGED4B through WB, and the results are shown in Figure 2. MAGED4B had the lowest protein expression in the STAD cell BGC and the highest protein expression in HGC-27. Therefore, we selected the BGC cells to construct the cell lines with overexpressed MAGED4B, and chose the HGC-27 cells to construct with cell lines MAGED4B knockdown (KD) for the subsequent cytological experiments.
 |
Figure 2 Expressions of MAGED4B in different STAD cell lines. |
MAGED4B Promotes the Invasion and Proliferation of Stomach Adenocarcinoma Cells
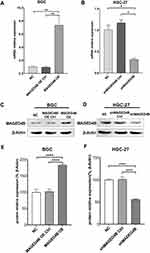
The qPCR and WB were adopted to verify whether the cell lines with overexpressed MAGED4B and the cell lines with silent MAGED4B were constructed successfully. The qPCR results showed that the expression of MAGED4B in the MAGED4B OE (MAGED4B overexpression) cell lines was significantly higher than that in the BGC cells and OE Ctrl (overexpression control) (P<0.01) (Figure 3A); the expression of MAGED4B in the MAGED4B KD (MAGED4B knockdown) cell lines was significantly lower than that in the BGC cells and OE Ctrl (P<0.01) (Figure 3B). The WB detection results of the MAGED4B protein expression were the same as that of qPCR. The protein expression of MAGED4B in the MAGED4B OE cell lines was significantly higher than that in the BGC cells and OE Ctrl (P<0.05) (Figure 3C); the protein expression of MAGED4B in the MAGED4B KD cell line was significantly lower than that in the BGC cells and OE Ctrl (P<0.05) (Figure 3D). The statistical results of protein expression are shown in Figure 3E and F.
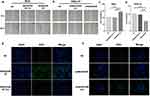
The migration ability of the MAGED4B OE and MAGED4B KD cells was evaluated in the single-layer wound healing experiment. The results showed that the open wound closure speed of cells in the MAGED4B OE group was significantly faster than that in the BGC and OE Ctrl groups (Figure 4A); on the contrary, the open wound closure speed of cells in the MAGED4B KD group was significantly slower than that in the HCG-27 and KD Ctrl (knockdown control) groups (Figure 4B); the above results proved that the overexpression of MAGED4B improved the migration ability of STAD cells. The cell viability was determined by the CCK-8 assay. The results showed that the cell viability of the MAGED4B OE group was significantly higher than that of the BGC and OE Ctrl groups (Figure 4C); on the contrary, the cell viability of the MAGED4B KD group was significantly lower than that of the HCG-27 and KD Ctrl groups (Figure 4D); the above results indicate that the overexpression of MAGED4B increased the cell viability of STAD cells. The proliferation ability of cells was determined by the EDU test. The results showed that the number of proliferated cells in the MAGED4B OE group was significantly higher than that in BGC and OE Ctrl groups (Figure 4E); on the contrary, the number of new cells in the MAGED4B KD group was significantly lower than that in the HCG-27 and KD Ctrl groups (Figure 4F); the above results proved that the overexpression of MAGED4B promoted the proliferation of STAD cells. To sum up, the overexpression of MAGED4B promotes the migration, viability and proliferation of stomach adenocarcinoma cells, while the silent MAGED4B inhibits the migration, viability and proliferation of stomach adenocarcinoma cells. These data indicate that MAGED4B is a key regulator for the migration, viability and proliferation of stomach adenocarcinoma cells.
MAGED4B Inhibits Apoptosis of Stomach Adenocarcinoma Cells and Their Sensitivity to Cisplatin
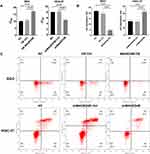
Cisplatin is a chemotherapeutic drug widely used in treatment of human solid malignant tumors. We measured the viability of STAD cells using the CCK8 method, so as to detect the effect of MAGED4B on the chemosensitivity of cisplatin. The results showed that the overexpression of MAGED4B (BGC) increased the IC50 of cisplatin, which means it decreased the sensitivity of STAD cells to cisplatin. On the contrary, the MAGED4B knockout (HGC-27) reduced IC50, that is, it increased the sensitivity of STAD cells to cisplatin (Figure 5A).
Then, we further examined the effect of MAGED4B on the sensitivity of STAD cells to cisplatin by flow cytometry. The apoptosis after incubation using cisplatin (8 µ g/mL) for 24 hours is shown in the Figure 5B and C. The knockdown of MAGED4B can promote the apoptosis of HGC-27 cells induced by cisplatin, while the overexpression of MAGED4B can inhibit the apoptosis of BGC cells induced by cisplatin. The above results indicate that MAGED4B reduces the sensitivity of STAD cells to cisplatin (Figure 5B and C).
MAGED4B Inhibits TNF-α Induced Apoptosis by Inhibiting TRIM27
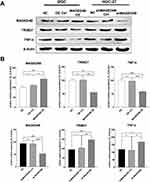
According to previous studies, the MAGED4B protein can interact with the RING finger protein to regulate tumor progression. As an E3 ubiquitin ligase with the RING finger domain, TRIM27 is proved to interact with MAGED4B.17 Therefore, we detected the expressions of TRIM27 in cells with overexpressed and knockdown MAGED4B by WB. The results are shown in Figure 6: The overexpression of MAGED4B down-regulated the protein expression of TRIM27, while the MAGED4B knockdown up-regulated the protein expression of TRIM27. Previous studies prove18 that TRIM27 can form a complex with ubiquitin specific protease USP7, and ubiquitinate USP7 to promote the TNF-α induced apoptosis. Therefore, we further evaluated the expression of TNF-α in the cells with overexpressed and knockdown MAGED4B, and found that the overexpression of MAGED4B down-regulated the protein expression of TNF-α, while the MAGED4B knockdown up-regulated the protein expression of TNF-α. Combined with the detection results of apoptosis by flow cytometry, we can speculate that the overexpression of MAGED4B inhibited the expression of TRIM27, thereby inhibiting the TNF-α induced apoptosis.
Discussion
The MAGED4B gene is a type II MAGE gene with unique distribution in normal tissues, which has specific expression in brain and ovary, and a high expression level in various malignant tumors.19–21 According to recent studies, MAGED4B has highly expressions in a variety of cancers, such as oracle square cell carcinoma, square cell carcinoma of the esophagus, and breast cancer.13,22,23 In this work, we used the TCGA stomach adenocarcinoma database to analyze the expression levels of MAGED4B in multiple stomach adenocarcinoma samples, and found that MAGED4B has shown continuous overexpression in the STAD tissues, indicating its carcinogenic effect in stomach adenocarcinoma.
In some recent studies, the high expression of MAGED4B in some primary cancers is associated with tumor progression and poor prognosis.22 For example, according to the study of Oya et al, the patients with high expression of MAGED4 mRNA in the esophageal cancer tissues had significantly shorter overall survival than the patients with low expression of MAGED4 mRNA (their 2-year survival rates were 44% and 73%, respectively, P=0.006). At the same time, the univariate and multivariate analysis results showed that the high expression of MAGED4 mRNA was an independent prognostic factor of the squamous cell carcinoma of the esophagus.22 Consistent with these findings, we found that according to the stomach adenocarcinoma data in TCGA, the high expression of MAGED4B was positively correlated with the low PFS in stomach adenocarcinoma patients.
Among all cancer biomarkers, some can play a functional role in tumor biology, while others have no contribution to the formation and progress of tumors due to the expression disorder, which may be just incidental in the malignant transformation. Therefore, the selection and characterization of molecular mechanism that plays a critical role in tumor behavior may help identify new targeted therapies.24 We conducted in vitro experiments on the STAD cells to study the effects of MAGE-D4B. The qRT-PCR results showed that MAGED4B was highly expressed in a variety of STAD cells, which was consistent with the results of tumor samples in vivo. By constructing the cell lines with overexpressed MAGED4B, we found that the overexpression of MAGED4B promoted the migration, viability and proliferation of stomach adenocarcinoma cells, while these functions were inhibited in the cell lines with silent MAGE-D4B. The effects of MAGED4B on the migration, viability and proliferation of STAD cells indicate that this protein plays a functional role in the tumorigenesis of stomach adenocarcinoma. Germano et al25 proved that the tumorigenic potential of MAGE-D4B may be attributed to the down-regulation of tumor suppressor E-cadherin, which plays a key role in maintaining the cell-cell adhesion.26,27
Our research results show that the overexpression of MAGED4B could reduce the sensitivity of the STAD cells to cisplatin, increase their IC50, and inhibit the cisplatin induced apoptosis. The WB results show that the overexpression of MAGED4B could down-regulate TRIM27 and TNF-α. TNF-α is an effective inflammatory cytokine that can induce apoptosis of tumor cells, but not that of normal cells.28 Recent studies show that ubiquitination plays a crucial role in the signal transmission of TNF-α.29 TRIM27 is a tripartite motif (TRIM) protein containing RING finger, B-box, and cooked coil domains, which can positively regulate TNF-α induced apoptosis.18 Related researches show that the Trim27 deficient mice presented drug resistance to the TNF-α-d-galactosamine induced apoptosis of hepatocytes. TRIM27 can form a complex with the ubiquitin specific protease USP7 and ubiquitinate it. USP7 can deubiquitinate the receptor interacting protein 1 (RIP1), thereby promoting the TNF-α induced apoptosis. The ubiquitination-deubiquitination cascade reaction mediated by the TRIM27-USP7 complex plays an important role in the TNF-α induced apoptosis.18 Therefore, we speculate that MAGED4B affects its ubiquitination of USP7 and the formation of complex by inhibiting TRIM27, so as to inhibit the downstream cascade reaction and finally inhibit the TNF-α induced apoptosis. However, the specific mechanism behind it still needs further exploration. In the future, we will continue to explore the molecular mechanism of how MAGED4B affects the functions of tumor cells, and find valuable prognostic biomarkers and attractive cancer treatment targets for treatment of the stomach adenocarcinoma.
Data sharing Statement
All relevant data are available from the corresponding authors on request.
Ethical Approval and Consent to Participate
This research was approved by the Ethics Committee of the Henan Province Hospital of Traditional Chinese Medicine. Written informed consent was obtained from the patients for the usage and publication of their clinical data. All research complied with The Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors have no relevant financial or non-financial interests to disclose.
References
1. Tan P, Yeoh KG. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149(5):1153–1162 e1153.
2. Zhang M, Hu S, Min M, et al. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut. 2021;70(3):464–475. doi:10.1136/gutjnl-2019-320368
3. Lochhead P, El-Omar EM. Gastric cancer. Br Med Bull. 2008;85:87–100. doi:10.1093/bmb/ldn007
4. Maruyama K, Kaminishi M; Japanese Gastric Cancer Association Registration C. Gastric cancer treated in 1991 in Japan: data analysis of nationwide registry. Gastric Cancer. 2006;9(2):51–66. doi:10.1007/s10120-006-0370-y
5. Hundahl SA, Menck HR, Mansour EG, Winchester DP. The national cancer data base report on gastric carcinoma. Cancer. 1997;80(12):2333–2341. doi:10.1002/(SICI)1097-0142(19971215)80:12<2333::AID-CNCR15>3.0.CO;2-V
6. Serlin O, Keehn RJ, Higgins GA
7. Adachi Y, Yasuda K, Inomata M, Sato K, Shiraishi N, Kitano S. Pathology and prognosis of gastric carcinoma: well versus poorly differentiated type. Cancer. 2000;89(7):1418–1424. doi:10.1002/1097-0142(20001001)89:7<1418::AID-CNCR2>3.0.CO;2-A
8. Siewert JR, Bottcher K, Stein HJ, Roder JD. Relevant prognostic factors in gastric cancer: ten-year results of the German gastric cancer study. Ann Surg. 1998;228(4):449–461. doi:10.1097/00000658-199810000-00002
9. Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67(6):705–712. doi:10.1002/jnr.10160
10. De Plaen E, Arden K, Traversari C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40(5):360–369. doi:10.1007/BF01246677
11. Jungbluth AA, Stockert E, Chen YT, et al. Monoclonal antibody MA454 reveals a heterogeneous expression pattern of MAGE-1 antigen in formalin-fixed paraffin embedded lung tumours. Br J Cancer. 2000;83(4):493–497. doi:10.1054/bjoc.2000.1291
12. van der Bruggen P, Traversari C, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi:10.1126/science.1840703
13. Chong CE, Lim KP, Gan CP, et al. Over-expression of MAGED4B increases cell migration and growth in oral squamous cell carcinoma and is associated with poor disease outcome. Cancer Lett. 2012;321(1):18–26. doi:10.1016/j.canlet.2012.03.025
14. Yang P, Huo Z, Liao H, Zhou Q. Cancer/testis antigens trigger epithelial-mesenchymal transition and genesis of cancer stem-like cells. Curr Pharm Des. 2015;21(10):1292–1300. doi:10.2174/1381612821666141211154707
15. Zhu W, Li H, Yu Y, et al. Enolase-1 serves as a biomarker of diagnosis and prognosis in hepatocellular carcinoma patients. Cancer Manag Res. 2018;10:5735–5745. doi:10.2147/CMAR.S182183
16. Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. J Vis Exp. 2014;88:e51046.
17. Liu C, Liu J, Shao J, et al. MAGED4B promotes glioma progression via inactivation of the tnf-alpha-induced apoptotic pathway by down-regulating TRIM27 expression. Neurosci Bull. 2022;39:273–291.
18. Zaman MM, Nomura T, Takagi T, et al. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2013;33(24):4971–4984. doi:10.1128/MCB.00465-13
19. Sasaki M, Nakahira K, Kawano Y, et al. MAGE-E1, a new member of the melanoma-associated antigen gene family and its expression in human glioma. Cancer Res. 2001;61(12):4809–4814.
20. Tsai JR, Chong IW, Chen YH, et al. Differential expression profile of MAGE family in non-small-cell lung cancer. Lung Cancer. 2007;56(2):185–192. doi:10.1016/j.lungcan.2006.12.004
21. Cheong SC, Chandramouli GV, Saleh A, et al. Gene expression in human oral squamous cell carcinoma is influenced by risk factor exposure. Oral Oncol. 2009;45(8):712–719. doi:10.1016/j.oraloncology.2008.11.002
22. Oya H, Kanda M, Takami H, et al. Overexpression of melanoma-associated antigen D4 is an independent prognostic factor in squamous cell carcinoma of the esophagus. Dis Esophagus. 2015;28(2):188–195. doi:10.1111/dote.12156
23. Fonseca-Sanchez MA, Perez-Plasencia C, Fernandez-Retana J, et al. microRNA-18b is upregulated in breast cancer and modulates genes involved in cell migration. Oncol Rep. 2013;30(5):2399–2410. doi:10.3892/or.2013.2691
24. Clynes M, O’Connor R, O’Driscoll L, Daly C, Meleady P. Challenges in molecular analysis for individualized cancer therapy. Drug Discov Today. 2003;8(12):531. doi:10.1016/S1359-6446(03)02730-2
25. Germano S, Kennedy S, Rani S, et al. MAGE-D4B is a novel marker of poor prognosis and potential therapeutic target involved in breast cancer tumorigenesis. Int J Cancer. 2012;130(9):1991–2002. doi:10.1002/ijc.26200
26. Baranwal S, Alahari SK. Molecular mechanisms controlling E-cadherin expression in breast cancer. Biochem Biophys Res Commun. 2009;384(1):6–11. doi:10.1016/j.bbrc.2009.04.051
27. Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3(5):289–293. doi:10.1186/bcr309
28. Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296(5573):1634–1635. doi:10.1126/science.1071924
29. Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12(7):439–452. doi:10.1038/nrm3143
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.