Back to Journals » OncoTargets and Therapy » Volume 11
Lupeol inhibits growth and migration in two human colorectal cancer cell lines by suppression of Wnt–β-catenin pathway
Authors Wang YH, Hong D, Qian YQ, Tu XZ , Wang KK, Yang XH, Shao SJ, Kong XL , Lou ZF, Jin LJ
Received 15 August 2018
Accepted for publication 26 September 2018
Published 9 November 2018 Volume 2018:11 Pages 7987—7999
DOI https://doi.org/10.2147/OTT.S183925
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper has been retracted.
Yihao Wang, 1,2,* Dan Hong, 1,* Yuqin Qian, 3 Xuezi Tu, 1 Keke Wang, 1 Xianhong Yang, 1 Sijia Shao, 1 Xinlong Kong, 1 Zhefeng Lou, 1 Longjin Jin 1
1School of Laboratory Medicine and Life Science, Wenzhou Medical University, Zhejiang, People’s Republic of China; 2School of Ophthalmology and Optometry, Wenzhou Medical University, Zhejiang, People’s Republic of China; 3School of the first Clinical Medical Sciences, Wenzhou Medical University, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Background: Lupeol, a triterpene isolated from various herbal plants, possesses an anti-inflammatory function and has been proposed as a candidate for anticancer agents. The purpose of this research was to investigate the effect of lupeol on the viability, apoptosis, cell-cycle distribution, and migration of colorectal cancer cell lines and its molecular mechanism.
Methods: Lupeol was assessed for its anticancer effect using two human colorectal cancer cell lines: SW480 and HCT116. These cells were treated with lupeol, and their viability, apoptosis, migration, and cycle distribution were detected by CCK8, flow cytometry, and the transwell method. Quantitative PCR, Western blot, and immunofluorescence were applied to detect the expressions of CTNNB1, TCF4, cMYC, CCND1, CLDN1, and CCNA2.
Results: Lupeol suppressed cell viability and migration and induced cellular apoptosis of both cell lines, with increased p53 and decreased Bcl2 protein levels (P< 0.05). Cell cycles of both lupeol-treated cell lines were arrested in the S phase (P< 0.05). Quantitative PCR and Western blot analyses showed significantly reduced expressions of CTNNB1, TCF4, and downstream genes of the Wnt–β-catenin pathway, including the cell-cycle-regulated genes of cMYC and CCND1 of both cell lines upon lupeol treatment (P< 0.05). mRNA and protein levels of CLDN1 decreased in HCT116 cells, plus the expression of CCNA2 mRNA and protein decreased in SW480 cells (P< 0.05). Immunofluorescence analysis confirmed decreased expression of Wnt–β-catenin signaling.
Conclusion: Our findings indicate that lupeol effectively inhibits proliferation and migration and induces apoptosis and cell-cycle arrest of two colorectal cell lines by inactivation of the Wnt–β-catenin signaling pathway and downregulation of cMYC, CCND1, CCNA2, and CLDN1, thereby making it a promising anticancer candidate.
Keywords: lupeol, colorectal cancer, Wnt, β-catenin signaling pathway, proliferation, apoptosis, migration
Introduction
Colorectal cancer (CRC) is the third-most common type of cancer in the world.1 Alkylating cytotoxic agents like oxaliplatin are usually combined with radiotherapy to treat stage II and stage III CRC, which confronts obstacles like multidrug resistance and severe side effects. These therapies ultimately lead to drug intolerance and tumor relapse in CRC patients.2 Plants contain a wide range of potential anticancer-drug substances with multifarious functions and targets. In recent years, triterpenes, including lupane, oleanane, and ursane, have shown their potential as anticancer candidates by various methods of administration, such as gavage and subcutaneous and intravenous injection in animal models.3
Lupeol is a pentacyclic triterpene in the lupane group, widely found in herbal plants like kale, mango, and dandelion, with anti-inflammatory, antioxidation, anti-infective, antihyperglycemia, antiasthma, antiarthritis, cardioprotective, neuroprotective, hepatoprotective, and chemosensitization effects.4,5 This evidence supports lupeol possessing varied pharmacological potency with many potential mechanisms and targets. However, its complicated molecular mechanism and whether lupeol might be a promising leading compound to treat CRC remain unclear.
The high prevalence of CRC calls for therapeutic agents targeting the mechanism of its premalignant lesions and evolution of cancer. An aberrantly activated Wnt–β-catenin signaling pathway is a common feature involved in 93% of CRC. Two most frequent gene mutations in carcinogenesis resulting in excessive accumulation of β-catenin are APC mutations (82%) and the activation of CTNNB1 itself (4.7%).6 These mutations lead to disruption of β-catenin degradation in cytoplasm. Excessive β-catenin translocates to the nucleus to bind with TCF/LEF transcription factors to form the β-catenin–TCF complex, which binds to promoter regions of CCND1 and cMYC, thereby accelerating metabolic activation of the cell cycle in malignant transformation.7 GeneCards and Kyoto Encyclopedia of Genes and Genomes signaling pathways both indicate that CCNA2 is located downstream of the Wnt pathway and regulated by Wnt-target gene transcription of cMyc.8 Overexpression of CCNA2 can result in delayed onset of cell division in mammalian cells.9 CCNA2 may also regulate oncoproteins or tumor suppressors like TP53, contributing to tumorigenesis.10 Also, CLDN1 is regulated by β-catenin and TCF4 and highly increased in CRC, helping promote cellular malignant transformation through regulating epithelial–mesenchymal transition (EMT).11 Therefore, targeting the cascade activation of Wnt–β-catenin signaling transcription is crucial for CRC therapy.12
Here, we explored the effects of lupeol on the viability, apoptosis, cell cycle, and migration of CRC cell lines, ie, SW480 (APC deleted, β-catenin wild type) and HCT116 (APC wild type, β-catenin mutant). Furthermore, we explored the mechanism of lupeol-mediated suppression in CRC cell lines of Wnt–β-catenin signaling by evaluating expressions of CTNNB1, TCF4, and Wnt–β-catenin signaling downstream genes cMYC, CCND1, CLDN1, and CCNA2.
Methods
Cell culture
The human CRC cell lines SW480 and HCT116 were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). Both cell lines are regularly authenticated on the basis of viability, recovery, growth, morphology, and chemical response, most recently confirmed 3–4 months before use. Both cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS with 1% penicillin–streptomycin (CellGro; Corning Incorporated, Corning, NY, USA) in humidified air with 5% CO2 at 37°C.
Chemical reagents
Lupeol was purchased from Sigma-Aldrich Co. (St Louis, MO, USA) and dissolved in dimethyl sulfoxide (DMSO) at a concentration of 2.4×10−2 mol/L as a stock solution. The solution was diluted to give a final DMSO concentration of 0.1% (v:v) and stored at −80°C. The control groups contained DMSO at this concentration. Cyclin A2 (ab38) and claudin 1 (ab15098) antibodies were purchased from Abcam (Cambridge, UK). β-Catenin (AF0066) and cyclin D1 (AF853) antibodies were bought from Beyotime (Haimen, People’s Republic of China). GAPDH (sc47724), cMyc (sc40), and β-actin (sc8432) antibodies were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). TCF4 (2565s), p53 (9282), Bcl2 (2872), antirabbit IgG (H+L, 4412), and antimouse IgG (H+L, 8890) antibodies were bought from Cell Signaling Technology (Danvers, MA, USA). Antirabbit (sc2004) and antimouse (sc2005) IgG–HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology Inc. Trizol reagent was bought from Thermo Fisher Scientific (Waltham, MA, USA).
Cell-viability assay
Cell viability was determined using CCK8 (Beyotime, Shanghai, People’s Republic of China). Cells (104 per well) were seeded into a 96-well plate in 100 μL RPMI 1640 supplemented with 10% FBS and allowed to adhere overnight. After treatment with lupeol at various concentrations for 24 and 48 hours, 10 μL CCK8 solution was added to each well of the plate. Plates were incubated at 37°C for 1 hour, after which absorbance at 450 nm was measured using a microplate reader (BioTek, Winooski, VT, USA). All experiments were performed in triplicate and repeated at least three times.
Cell-apoptosis assay
Cells (2×105) were stained with fluorescein isothiocyanate-conjugated anti-annexin V antibody, which was labeled in combination with propidium iodide (PI) according to the manufacturer’s protocol (KeyGen Biotech, Nanjing, People’s Republic of China), then analyzed by fluorescence-activated cell sorting (BD, Franklin Lakes, NJ, USA). The cell-death percentage corresponded to annexin V+–PI+ cells. The apoptotic cell-death percentage was represented by annexin V+–PI−-stained cells.
Cell-cycle analysis
Cells (2×105) were cultured in each well of six-well plates till 60% confluent with normal culture medium. Cells were synchronized by replacing the medium containing 0.1% FBS for 12 hours to arrest them in the G0 phase of the cell cycle, after which they were treated with or without lupeol in RPMI 1640 complete media for 24 hours. Cells were trypsinized thereafter, washed twice with cold PBS, and centrifuged at 1,500 rpm. For cell-cycle analysis, cells were fixed in prechilled 70% ethanol overnight at 4°C. Cells were centrifuged at 1,500 rpm for 5 minutes, pellets washed twice with cold PBS, suspended in 500 μL PBS, and incubated with 5 μL RNase (Takara, Kusatsu, Japan) at 37°C for 30 minutes. Cells were chilled over ice for 10 minutes and stained with PI staining solution (PI 50 mg/mL, RNase A 10 mg/mL, and 0.1% Triton X-100; Sigma-Aldrich Co.) for 1 hour and analyzed by FlowJo version 10 flow cytometry.
Cell-migration assay
In vitro cell migration was determined following the manufacturer’s instructions. Cells (5×104) were seeded in each chamber and left for 24 hours. The stoppers used to create the migration zone were removed after 24 hours, and cells washed with PBS twice to remove any unattached cells. Fresh RPMI 1640 complete medium (100 μL) with lupeol or DMSO was added to each well. Cells were allowed to migrate into the migration zone. After incubation with the transwell assay for 48 hours, cells on both the inside and outside of the seeded plate were fixed with 4% formalin, then transferred to 100% methanol, and finally stained with crystal violet at room temperature in the dark. They were washed again, then noncrossing cells were scraped off with a cotton swab (PBS-wetted). Cells were fluorescently stained with CellTracker green (Thermo Fisher Scientific). Fluorescent signals were measured using a microplate reader (Synergy HT; BioTek) with 492 nm excitation and 517 nm emission filters.
Reverse-transcription and real-time polymerase chain reaction
In this reaction, 2 μg total RNA was reverse-transcribed with random primers and SuperScript IV reverse transcriptase according to the user’s manual (Thermo Fisher Scientific). The reaction was performed with incubation at 42°C for 1 hour, and the enzymes were subsequently inactivated by incubation at 85°C for 5 minutes. cDNA was used for real-time PCR analysis with gene-specific primers, and gene expression was detected using a Fast SYBR green master mix (Thermo Fisher Scientific). RNA expression was normalized to that of GAPDH (ΔCt = target RNA Ct − GAPDH Ct). Individual primers used in this study were: CTNNB1-F: 5′-CATCTACACAGTTTGATGCTGCT-3′, R: 5′-GCAGTTTTGTCAGTTCAGGGA-3′; TCF4-F: 5′-GGCTATGCAGGAATGTTGGG-3′, R: 5′-GTTCATGTGGATGCAGGCTAC-3′; MYC-F: 5′-GTCAAGAGGCGAACACACAAC-3′, R: 5′-TTGGACGGACAGGATGTATGC-3′; CCND1-F: 5′-GCTGCGAAGTGGAAACCATC-3′, R: 5′-CCTCCTTCTGCACACATTTGAA-3′; CCNA2-F: 5′-GGATGGTAGTTTTGAGTCACCAC-3′, R: 5′-CACGAGGATAGCTCTCATACTGT-3′; CLDN1-F: 5′-AGCACCGGGCAGATACAGT-3′, R: 5′-GCCAATTACCATCAAGGCTCG-3′; and GAPDH-F: 5′-GGAGCGAGATCCCTCCAAAAT-3′, R: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western blot analysis
Briefly, CRC cells were washed three times after lupeol treatment for 24 hours in cold PBS and lysed in lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, and 1% Triton X-100 (Beyotime) supplemented with a protease-inhibitor cocktail and phosphatase inhibitor cocktail (MCE) for 30 minutes on ice. Homogenates were centrifuged at 12,000 rpm for 10 minutes, and extracted protein concentrations were measured using BCA assay (23227; Thermo Fisher Scientific) and determined by Bio-Rad protein assay. Protein lysates (~40 μg) were electrophoresed in SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes (ISEQ00010; EMD Millipore, Billerica, MA, USA) by electroblotting. The membranes were blocked with Tris-buffered saline with Tween 20 (TBST) containing 5% dried milk–BSA for 1 hour at room temperature, followed by incubation with primary antibody for 16 hours. Antibodies of β-catenin (1:8,000, rabbit), TCF4 (1:400, rabbit), cMyc (1:400, mouse), cyclin D1(1:147, mouse), β-actin (1:1,000, mouse), cyclin A2 (1:200, mouse), claudin 1 (1:200, rabbit), p53 (1:1,000, rabbit), Bcl2 (1:1,000, rabbit), and GAPDH (1:800, mouse) were incubated. Membranes were washed three times with TBST. Then, membranes were incubated with antirabbit (sc2004) or antimouse (sc2005) IgG–HRP-conjugated secondary antibodies (1:10,000) for 1 hour. After three washes with TBST (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.1% Tween 20), immunoreactive bands were detected using a WesternBright enhanced chemiluminescence (RPN2135; GE Healthcare UK Ltd., Little Chalfont, UK). GAPDH or β-actin was used as an internal loading control.
Immunofluorescence
Cells were seeded on aseptic 1 cm-diameter disks in a 24-well plate at a density of 2×104 cells/disk for 4 days. Samples were washed with PBS and fixed in 4% paraformaldehyde for 15 minutes at room temperature. Cells were washed with PBS three times and blocked with PBS containing 5% BSA for 1 hour. After being blocked with BSA, samples were incubated with primary antibody β-catenin (1:200, rabbit; Beyotime), TCF4 (1:50, rabbit; Cell Signaling Technology), cMyc (1:50, mouse; Santa Cruz Biotechnology), cyclin D1 (1:20, mouse; Beyotime), claudin 1 (1:80, rabbit; Abcam), and cyclin A2 (1:80, mouse; Abcam) overnight at 4°C. On the following day, samples were washed with PBS three times and incubated with goat antirabbit antibody (1:500) or goat antimouse antibody (1:500), which were diluted in a blocking solution for 1 hour at room temperature. Samples were further counterstained with DAPI (1:500; Boster, Pleasanton, CA, USA) and phalloidin (1:100; Beyotime). Inverted fluorescence microscopy (Eclipse Ti; Nikon Corporation, Tokyo, Japan) was used to capture fluorescent images.
Statistical analysis
Results were statistically analyzed using Student’s t-test with SPSS 22.0, which were analyzed using GraphPad Prism version 6.0. All experiments were independently repeated at least three times. P<0.05 was considered statistically significant. All data are represented as mean ± SD.
Results
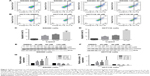
Effect of lupeol on viability of SW480 and HCT116 cells
We investigated the effect of lupeol (20–120 μM) treatment on the viability of an APC-deficient cell line (SW480) and β-catenin-mutated cell line (HCT116) by CCK8 assay. The viability of both cell types treated with lupeol was decreased in a dose-dependent manner at 24 and 48 hours’ lupeol treatment (Figure 1A and B). The IC50 of lupeol treatment at 24 hours of SW480 and HCT116 cells was 106.3 and 62.0 μM, respectively. The IC50 at 48 hours of SW480 and HCT116 cells was 90.2 and 53.3 μM, respectively. Based on cell-viability results, we selected various concentrations of lupeol (0, 40, 80, and 120 μM) for SW480 cells and HCT116 cells (0, 20, 40, and 80 μM) with 24 hours’ treatment for subsequent studies.
Effect of lupeol on apoptosis of SW480 and HCT116 cells
In the cellular apoptosis study, both cell lines were treated with lupeol or DMSO and assessed by flow cytometry with annexin V–PI double staining. Results showed that lupeol induced apoptosis in SW480 and HCT116 cells in a dose-dependent manner (P<0.05; Figure 2A–D). The p53-wild type HCT116 cells exerted higher apoptosis induction to lupeol exposure than p53-mutated SW480 cells. To explore the molecular mechanism of lupeol in CRC-cell apoptosis, we detected the Bcl2 and p53 protein levels assessed by Western blot (Figure 2E and F). The p53 protein level was increased in lupeol-treated groups. Conversely, Bcl2 level was inhibited by high lupeol concentration (Figure 2G and H). Our results indicated that lupeol promoted apoptosis in CRC cells via the p53 pathway.
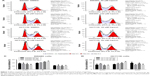
Effect of lupeol on cell cycle in SW480 and HCT116 cells
Next, we considered the possibility that growth inhibition of CRC cells may involve an arrest of cells at specific check points in the cell cycle followed by apoptosis. Therefore, we assessed the effect of lupeol on cell-cycle perturbation (Figure 3A and B). Lupeol treatment significantly increased the number of SW480 and HCT116 cells in the S phase of the cell cycle (Figure 3C and D). This suggested that lupeol significantly induced S-phase cell-cycle arrest of both CRC cell types.
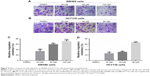
Effect of lupeol on cell migration in SW480 and HCT116 cells
Cell-migration assays were applied using the transwell method. Both CRC cell types exhibited remarkably reduced migration after lupeol treatment (Figure 4A and B). Results showed that 40 and 80 μM lupeol almost absolutely blocked cell invasion for HCT116 and SW480 cells, respectively. In sum, lupeol significantly inhibited migration of SW480 and HCT116 cells in a dosage-dependent manner (Figure 4C and D).
Effect of lupeol on transcription activity in Wnt–β-catenin pathway
We next evaluated the effect of lupeol treatment on transcription levels in the Wnt–β-catenin pathway for SW480 and HCT116 cell lines using quantitative reverse-transcription PCR. Lupeol treatment caused a significant decrease in the transcription level of CTNNB1 expression in HCT116 cells, while not causing significant change in SW480 cells. Lupeol treatment also inhibited cMYC- and CCND1-transcription activation in SW480 and HCT116 cells (P<0.05). Interestingly, transcription levels of TCF4 and CLDN1 in HCT116 cells were decreased and CCNA2 mRNA in SW480 cells decreased to different degrees, also exhibiting significant differences compared to the control group (P<0.05; Figure 5A and B).
Effect of lupeol on the protein levels in Wnt–β-catenin pathway
Next, we sought to explore protein levels at which lupeol suppressed CRC cell lines. We detected protein-expression levels of β-catenin, TCF4, cMyc, cyclin D1, claudin 1, and cyclin A2 in SW480 and HCT116 cells by Western blot analysis (Figure 6A–D). Results showed that lupeol decreased β-catenin and TCF4 mRNA expression in a dosage-dependent manner (Figure 6E and F). In both lupeol-treated cell lines, protein-expression levels of cMyc and cyclin D1 decreased compared with the control group (P<0.05; Figure 6E and F). Corresponding with mRNA expression, protein-expression levels of claudin 1 in HCT116 cells and cyclin A2 in SW480 cells upon lupeol treatment decreased compared with the control group (P<0.05; Figure 6E and F). This finding suggests that lupeol attenuated Wnt–β-catenin signaling activity and suppressed the expression of cMyc, cyclin D1, and cyclin A2, thus impeding proliferation and arresting the cell cycle in CRC cells.
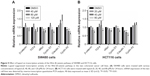
Effect of lupeol on protein expression in Wnt–β-catenin pathway
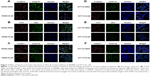
Translocation of β-catenin to the nucleus is vital for transcription of downstream Wnt–β-catenin genes. We explored the localization of β-catenin, TCF4, cMyc, cyclin D1, claudin 1, and cyclin A2 after lupeol treatment. In HCT116 cells, there was a decrease in β-catenin protein expression in lateral cell membranes after lupeol treatment (Figure 7B). However, no inhibited translocation of β-catenin to the nucleus was observed in SW480 cells (Figure 7A). We further observed decreased fluorescence intensities of cMyc and cyclin D1 in nuclei in both CRC cell lines (Figure 7A, B, D, and E). Corresponding with Western blot analysis, fluorescence intensity of claudin 1 protein in HCT116 cells and cyclin A2 in SW480 cells was decreased upon lupeol treatment compared with control group (Figure 7C and F).
Discussion
Epidemiological evidence has shown an association between triterpenoid-enriched plant intake and decreased risk of developing CRC.13 Likewise, considerable evidence has shown that triterpenoids, such as lupeol, show an antitumor effect in certain cell lines.14,15 DNA damage is identified as a premise of cancer development, and lupeol has the potentiality to inhibit chemically induced DNA damage, both in vitro and in vivo.16 Recent studies have shown that in addition to chemoprevention, lupeol has a chemotherapeutic effect on prostate cancer, hepatic carcinoma, and gallbladder cancer.14,17–19 Lupeol can also regulate the immunoresponse and impart chemotherapeutic resistance.15,20 In a previous study, lupeol showed inhibition of melanoma cells with abnormally active Wnt–β-catenin signals by reducing Wnt signaling.21 Therefore, we wanted to evaluate the effect of lupeol treatment on different CRC cells with abnormally activated Wnt–β-catenin pathways. In the present study, we used two variants of CRC cell lines: SW480 cells (APC deleted, β-catenin wild type) and HCT116 cells (APC wild type, β-catenin mutated). Our results showed that 20–120 μM lupeol treatment for 24 hours resulted in decreased cell viability and increased cellular apoptosis in a dose-dependent manner. These results are in accord with earlier research that also indicated that lupeol had no significant inhibitory effect on CRC cells that did not harbor abnormal activation of Wnt–β-catenin signaling.22 We further discovered that lupeol arrested the cell cycle at the S phase and reduced the cell-migration capability of the two cell lines compared to the control groups. Molecular studies revealed that the anticancer effect of lupeol may in part be mediated by Wnt–β-catenin signaling-pathway suppression.
Constitutive Wnt–β-catenin pathway activation is characteristic of most CRC tumorigenesis.6 Wnt–β-catenin pathway plays a pivotal role in regulating homeostasis and self-renewal of tissue in which β-catenin is involved in regulation of cell adhesion and gene transcription.23–25 β-Catenin in the nucleus serves to activate TCF-dependent transcription, leading to increased expression of downstream genes, including cMYC, CCND1, CCNA2, and MMP7.7,9,26 Studies have shown that β-catenin also interacts with growth-factor receptors in signal-transduction patterns during tumorigenesis.27 Tarapore et al found that lupeol significantly reduced CTNNB1-transcription levels in DLD and HCT116 cells with abnormally activated Wnt–β-catenin signals assessed by TopFlash assay, which supports the inhibitory action of lupeol on Wnt–β-catenin signaling.22 In our study, lupeol treatment decreased expression of β-catenin and TCF4 protein and decreased mRNA and protein expression of downstream cMYC and CCND1 in two CRC cell lines. The inhibitory effect of lupeol on highly metastatic HCT116 cells was stronger than that on SW480 cells, and the expression of β-catenin in HCT116 cells on lateral cell membranes associated with cell attachment was suppressed.28 In SW480 cells, inhibition of β-catenin translocation was not observed after lupeol treatment, which might have been due to subsequent ubiquitination and degradation of β-catenin, which could affect its positive control over transcription activity in the nucleus.29 Therefore, it is possible that the anticancer effect of lupeol in CRC cells is due to reduced nuclear expression of CTNNB1 and formation of β-catenin–TCF4 complexes, with subsequent disruption of signal transduction in the Wnt–β-catenin pathway. Furthermore, lupeol treatment resulted in significant decreases in the viability and migration of SW480, HCT116, and DLD1 cells with APC or CTNNB1 mutations, with no effect on RKO cells carrying wild-type APC and CTNNB1.22 This indicates that the inhibitory effect of lupeol on cell proliferation is probably more sensitive to CRC cells with APC and CTNNB1 mutations.
Several β-catenin/TCF4 target genes like cMYC and CCND1 are supposed to accelerate metabolic activation of the cell cycle. cMyc interacts with prereplication to form a complex located in the early-DNA-synthesis site, which has a direct impact on DNA replication. Its overexpression bypasses the G1/S phase-division checkpoint, increasing DNA-replication activity and DNA damage.30 Cyclin D–CDK4/6 complexes block the transcription of genes, negatively controlling cell cycles like that of the Rb tumor suppressor protein and allow the cell to go through the G1 checkpoint, thereby regulating cell-cycle progression and sustaining genomic integrity.31 Cyclin A2 is synthesized at the beginning of the S phase and binds to CDK2 to promote DNA synthesis.32 In our study, lupeol significantly reduced cell viability, induced apoptosis, and blocked the cell cycle in the S phase of the two CRC cell lines. Furthermore, quantitative PCR and Western blot analyses showed mRNA and protein expression of downstream CCND1 and cMYC was reduced. CCNA2 was downregulated in SW480 cells, but not in HCT116 cells. Similarly, lupeol can arrest the cell cycle in the S phase by reducing the expression of β-catenin protein and CCND1 and cMYC transcription in hepatoma and melanoma cells.33,34 Since the synthesis of DNA, histones, and related enzymes take place in the S phase, it is suggested that lupeol could reduce protein levels of β-catenin and TCF4 and reduce mRNA and protein expression of downstream cycle genes like cMYC and CCND1 in both cell lines and CCNA2 in SW480 cells so as to inhibit cell proliferation and arrest the cell cycle of CRC cells by repressing tetraploid formation and thus the mitosis process.26
Cancer-cell invasion and migration are essential for cancer metastasis, and the claudin family is closely related to these processes. In tumorigenesis, excessive TCF4 binds claudin 1 to certain sites to promote CLDN1 transcription,35 which not only contributes to EMT36 but also increases the expression of matrix metalloproteinases, promotes extracellular matrix destruction and tumor infiltration,37 and increases myosin–actin contractility to promote cell invasion and migration without affecting cell proliferation.38,39 Our results showed that lupeol downregulated CLDN1 expression in HCT116 cells, corresponding to their reduced migration rate evaluated by transwell assay, suggesting decreased myosin–actin interaction-mediated cell motility. In addition, β-catenin–TCF4 downstream CCNA2 is a novel target in CRC.40 Downregulation of CCNA2 displays increased cell invasiveness by actin-filament redistribution through regulation of the Rho family, such as inactivation of RHOA, leads to a decrease in cell adhesion, and promotes EMT processes.41,42 Cyclin A–CDK2 regulates APC mitotic spindle anchoring by phosphorylation of APC, which restrains microtubule attachment in mitosis.43 While lupeol significantly inhibited migration of SW480 cells, however, CCNA2 instead of CLDN1 was downregulated. Although the results were not anticipated from our previous study on migration of SW480 cells, downregulation of CCNA2 did exist. Since CCNA2 regulates much progress in the genesis and development of CRC, like cell migration and the cell cycle, the increase or decrease in its expression in cancer cells cannot fully explain the invasion and migration of cells.27,42,44 Therefore, the specific mechanism needs to be studied further.
Conclusion
We have provided evidences of anti-CRC effect of lupeol in cell viability, apoptosis, migration, cell-cycle arrest, and inactivation of Wnt–β-catenin signaling activity with the intervention of β-catenin nuclear translocation. The in vivo anticancer effect of lupeol is still not completely understood. Further research will be needed to elucidate the complicated mechanism of lupeol-induced Wnt–β-catenin inactivation, such as the knockdown assay. In addition, as lupeol is a pentacyclic triterpenoid belonging to over 30 triterpenoids that should also be explored, our research can provide evidence of the importance of dietary triterpenoids and interaction of lupeol with frequently mutated genes in initiative carcinogenesis pathways of CRC.
Data sharing statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors would like to thank Dr Magid Nisar from Pakistan for editing the English text of a draft of this manuscript. In addition, YHW especially wishes to thank Professor Suhao Lin for his continuous guidance and support to her. This study was presented as an abstract at the Zhejiang Medical Genetics Annual Conference (2018, Hangzhou, People’s Republic of China) and supported by grants from the Zhejiang Provincial Natural Science Foundation of China (LY13H250002).
Author contributions
YHW, LJJ, ZFL, and DH conceived and designed the study. YHW, DH, YQQ, and XHY drafted the manuscript. YHW, DH, YQQ, XZT, and KKW participated in implementation of the study. XHY, SJS, and XLK assisted in collecting the data. DH, YQQ, and KKW performed the statistical analysis. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. | ||
Saunders M, Iveson T. Management of advanced colorectal cancer: state of the art. Br J Cancer. 2006;95(2):131–138. | ||
Laszczyk MN. Pentacyclic triterpenes of the lupane, oleanane and ursane group as tools in cancer therapy. Planta Med. 2009;75(15):1549–1560. | ||
Saleem M. Lupeol, a novel anti-inflammatory and anti-cancer dietary triterpene. Cancer Lett. 2009;285(2):109–115. | ||
Siddique HR, Saleem M. Beneficial health effects of lupeol triterpene: a review of preclinical studies. Life Sci. 2011;88(7–8):285–293. | ||
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. | ||
Shang S, Hua F, Hu ZW, Zw H. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–33989. | ||
Lal A, Navarro F, Maher CA, et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–625. | ||
den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153(1):121–136. | ||
Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376(6535):88–91. | ||
Bhat AA, Sharma A, Pope J, et al. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PLoS One. 2012;7(6):e37174. | ||
Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. | ||
Mcdougall GJ, Allwood JW, Pereira-Caro G, et al. Novel colon-available triterpenoids identified in raspberry fruits exhibit antigenotoxic activities in vitro. Mol Nutr Food Res. 2017;61(2):1600327. | ||
Prasad S, Nigam N, Kalra N, Shukla Y. Regulation of signaling pathways involved in lupeol induced inhibition of proliferation and induction of apoptosis in human prostate cancer cells. Mol Carcinog. 2008;47(12):916–924. | ||
Zhu Y, Li X, Chen J, et al. The pentacyclic triterpene lupeol switches M1 macrophages to M2 and ameliorates experimental inflammatory bowel disease. Int Immunopharmacol. 2016;30:74–84. | ||
Srivastava AK, Mishra S, Ali W, Shukla Y. Protective effects of lupeol against mancozeb-induced genotoxicity in cultured human lymphocytes. Phytomedicine. 2016;23(7):714–724. | ||
Saleem M, Murtaza I, Witkowsky O, Kohl AM, Maddodi N. Lupeol triterpene, a novel diet-based microtubule targeting agent: disrupts survivin/cFLIP activation in prostate cancer cells. Biochem Biophys Res Commun. 2009;388(3):576–582. | ||
Liu F, He Y, Liang Y, et al. PI3-kinase inhibition synergistically promoted the anti-tumor effect of lupeol in hepatocellular carcinoma. Cancer Cell Int. 2013;13(1):108–114. | ||
Liu Y, Bi T, Shen G, et al. Lupeol induces apoptosis and inhibits invasion in gallbladder carcinoma GBC-SD cells by suppression of EGFR/MMP-9 signaling pathway. Cytotechnology. 2016;68(1):123–133. | ||
Liu Y, Bi T, Dai W, et al. Lupeol enhances inhibitory effect of 5-fluorouracil on human gastric carcinoma cells. Naunyn Schmiedebergs Arch Pharmacol. 2016;389(5):477–484. | ||
Tarapore RS, Siddiqui IA, Saleem M, Adhami VM, Spiegelman VS, Mukhtar H. Specific targeting of Wnt/β-catenin signaling in human melanoma cells by a dietary triterpene lupeol. Carcinogenesis. 2010;31(10):1844–1853. | ||
Tarapore RS, Siddiqui IA, Adhami VM, Spiegelman VS, Mukhtar H. The dietary terpene lupeol targets colorectal cancer cells with constitutively active Wnt/β-catenin signaling. Mol Nutr Food Res. 2013;57(11):1950–1958. | ||
Yan M, Li G, An J. Discovery of small molecule inhibitors of the Wnt/β-catenin signaling pathway by targeting β-catenin/Tcf4 interactions. Exp Biol Med. 2017;242(11):1185–1197. | ||
Sebio A, Kahn M, Lenz HJ. The potential of targeting Wnt/β-catenin in colon cancer. Expert Opin Ther Targets. 2014;18(6):611–615. | ||
Deitrick J, Pruitt WM. Wnt/β catenin-mediated signaling commonly altered in colorectal cancer. Prog Mol Biol Transl Sci. 2016;144:49–68. | ||
Kolligs FT, Bommer G, Göke B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002;66(3):131–144. | ||
Saleem M, Murtaza I, Tarapore RS, et al. Lupeol inhibits proliferation of human prostate cancer cells by targeting beta-catenin signaling. Carcinogenesis. 2009;30(5):808–817. | ||
Kaler P, Augenlicht L, Klampfer L. Activating mutations in β-catenin in colon cancer cells alter their interaction with macrophages; the role of snail. PLoS One. 2012;7(9):e45462. | ||
Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–33989. | ||
Dorasamy MS, Choudhary B, Nellore K, Subramanya H, Wong PF. Dihydroorotate dehydrogenase inhibitors target c-Myc and arrest melanoma, myeloma and lymphoma cells at S-phase. J Cancer. 2017;8(15):3086–3098. | ||
Dozier C, Mazzolini L, Cénac C, et al. CyclinD-CDK4/6 complexes phosphorylate CDC25A and regulate its stability. Oncogene. 2017;36(26):3781–3788. | ||
Yan J, Hao C, Delucia M, et al. CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J Biol Chem. 2015;290(21):13279–13292. | ||
Saleem M, Maddodi N, Abu Zaid M, et al. Lupeol inhibits growth of highly aggressive human metastatic melanoma cells in vitro and in vivo by inducing apoptosis. Clin Cancer Res. 2008;14(7):2119–2127. | ||
He Y, Liu F, Zhang L, et al. Growth inhibition and apoptosis induced by lupeol, a dietary triterpene, in human hepatocellular carcinoma cells. Biol Pharm Bull. 2011;34(4):517–522. | ||
Ouban A. Claudin-1 role in colon cancer: an update and a review. Histol Histopathol. 2018;33(10):1013–1019. | ||
Zhao X, Zou Y, Gu Q, et al. Lentiviral vector mediated claudin1 silencing inhibits epithelial to mesenchymal transition in breast cancer cells. Viruses. 2015;7(6):2965–2979. | ||
Smith JJ, Deane NG, Dhawan P, Beauchamp RD. Regulation of metastasis in colorectal adenocarcinoma: a collision between development and tumor biology. Surgery. 2008;144(3):353–366. | ||
van Itallie CM, Tietgens AJ, Anderson JM. Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol Biol Cell. 2017;28(4):524–534. | ||
Smith JJ, Deane NG, Dhawan P, Beauchamp RD. Regulation of metastasis in colorectal adenocarcinoma: a collision between development and tumor biology. Surgery. 2008;144(3):353–366. | ||
Yang F, Hu Y, Liu HX, Wan YJ. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem. 2015;290(10):6507–6515. | ||
Arsic N, Bendris N, Peter M, et al. A novel function for cyclin A2: control of cell invasion via RhoA signaling. J Cell Biol. 2012;196(1):147–162. | ||
Cheung CT, Bendris N, Paul C, et al. Cyclin A2 modulates EMT via β-catenin and phospholipase C pathways. Carcinogenesis. 2015;36(8):914–924. | ||
Beamish H, de Boer L, Giles N, Stevens F, Oakes V, Gabrielli B. Cyclin A/cdk2 regulates adenomatous polyposis coli-dependent mitotic spindle anchoring. J Biol Chem. 2009;284(42):29015–29023. | ||
Yasmeen A, Berdel WE, Serve H, Müller-Tidow C. E- and A-type cyclins as markers for cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2003;3(5):617–633. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.