Back to Journals » Infection and Drug Resistance » Volume 12
Low prevalence of resistance genes in sheltered homeless population in Marseille, France, 2014–2018
Authors Ly TDA, Hadjadj L, Hoang VT , Louni M, Dao TL, Badiaga S, Tissot-Dupont H, Raoult D, Rolain JM , Gautret P
Received 18 January 2019
Accepted for publication 7 March 2019
Published 7 May 2019 Volume 2019:12 Pages 1139—1151
DOI https://doi.org/10.2147/IDR.S202048
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Tran Duc Anh Ly,1,2 Linda Hadjadj,2,3 Van Thuan Hoang,1–2,4 Meriem Louni,1,2 Thi Loi Dao,1–2,5 Sekene Badiaga,2,6 Herve Tissot-Dupont,2,3 Didier Raoult,2,3 Jean-Marc Rolain,2,3 Philippe Gautret1,2
1IRD, AP-HM, SSA, VITROME, Aix Marseille Univ., Marseille, France; 2IHU-Méditerranée Infection, Marseille, France; 3MEPHI, Aix Marseille Univ., Marseille, France; 4Family Medicine Department, Thai Binh University of Medicine and Pharmacy, Thành Phố Thái Bình, Vietnam; 5Pneumology Department, Thai Binh University of Medicine and Pharmacy, Thành Phố Thái Bình, Vietnam; 6Emergency Department, North Hospital, AP-HM, Marseille, France
Objectives: The present study has explored the prevalence and potential factors contributing to the presence of nasal/pharyngeal resistant genes in homeless people.
Methods: During the winters 2014–2018, we enrolled sheltered homeless adults and controls and collected nasal/pharyngeal samples. Sixteen antibiotic resistance genes (ARGs), including genes encoding for beta-lactamases and colistin-resistance genes, were searched by real-time polymerase chain reaction (qPCR) performed directly on respiratory samples and followed by conventional PCR and sequencing.
Results: Over a 5-year period, using qPCR, we identified in homeless group (n=715) the presence of blaTEM (396/710, 54.7%), blaSHV (27/708, 3.6%), blaOXA-23 (1/708, 0.1%), while other genes including colistin-resistance genes (mcr-1 to mcr-5) were absent. We found a significantly higher proportion of ARG carriage among controls (74.1%) compared to homeless population (57.1%), p=0.038. Tobacco smoking (OR=4.72, p<0.0001) and respiratory clinical signs (OR=4.03, p=0.002) were most prevalent in homeless people, while vaccination against influenza (OR=0.31, p=0.016) was lower compared to controls. Among homeless people, type of housing (shelter A versus B, OR=1.59, p=0.006) and smoking tobacco (smoker versus non-smoker, OR=0.55, p=0.001) were independent factors associated with ARG carriage. By sequencing, we obtained a high diversity of blaTEM and blaSHV in both populations.
Conclusion: The lower risk for ARGs in the homeless population could be explained by limited access to health care and subsequently reduced exposure to antibiotics.
Keywords: antibiotic resistance gene, homeless, real-time polymerase chain reaction (qPCR), potential risk factors
Introduction
Homelessness is an increasingly social and public health concern in both developing and developed countries. Because of poor environmental conditions, poor physical state and substance abuse, this population is associated with the transmission of communicable diseases, including respiratory tract infections and multidrug-resistant tuberculosis.1,2 Antimicrobial resistance, if occurring in the homeless population, can challenge local health care systems because of the lack of surveillance due to the high mobility of this population. Several studies are available regarding the prevalence of antimicrobial resistance in homeless populations. An 8–10% prevalence of methicillin-resistant Staphylococcus aureus was evidenced in nasal sample from homeless people in Kansas City and in Boston, USA with resistance to erythromycin, levofloxacin and clindamycin.3,4 A study conducted in New Orleans, USA evidenced that housing in homeless shelter was an independent risk factor for high level of penicillin-non-susceptible Streptococcus pneumoniae carriage.5
Resistance to antibiotics may be due to the production of specific inactivating enzymes, changes in membrane permeability and efflux of the antibiotic, or to alteration of target sites.6 The production of beta-lactamases (extended-spectrum β-lactamases [ESBLs], especially those belonging to the CTX-M family, carbapenem-hydrolyzing β-lactamases) in Gram-negative bacteria is considered the most important mechanism contributing to antimicrobial resistance to β-lactam antibiotics.7 Due to be plasmid-borne, β-lactamases might spread among aerobic Gram-negative bacilli including Enterobacteriaceae and non-lactose fermenting bacteria (Pseudomonas aeruginosa, Acinetobacter baumannii).7 Carbapenemases belonging to the blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-58 subgroups are those most frequently identified from A. baumannii.8 Multidrug-resistant A. baumannii which has been reported a frequent cause of respiratory infection might be responsible for nosocomial infection outbreaks and lead to increase health care-associated costs and occurrence of hard-to-treat bacterial infections.9,10 Colistin is currently regarded as one of the last-resource antibiotics for a variety of treatment of human infections.11 Nevertheless, the emergence of the plasmid-mediated colistin resistance genes, such as mcr-1 gene, has also been documented in Enterobacteriaceae species and then disseminated worldwide due to the spread of a transposon, which can move in or out of plasmids and the chromosome.11
The burden of antibiotic resistance in Marseille homeless population remains unknown and unexplored. Therefore, in this cross-sectional study, we aimed to assess the prevalence of most common resistance genes carriage (genes encoding for β-lactamases and colistin-resistance genes) in nasal-pharyngeal samples from sheltered homeless people in comparison with “non-homeless” controls. We also investigate the role of potential risk factors for resistance gene carriage. Rather than exploring antibiotic resistance by conventional culture-based methods, a molecular technique was directly applied, as this strategy has proven successful in other populations.12
Materials and methods
Selection of homeless participants
In our one-shot studies, adult homeless people residing in two municipal emergency shelters (shelters A and B) in Marseille, France were recruited on a voluntary basis in winter from 2014 through 2018. The participants were asked to complete questionnaires including information on demographics, personal history, substance use, vaccination status and respiratory clinical presentation at the time of enrolment.
Selection of comparison group
Controls (defined as the non-homeless group) included administrative staffs, physicians, nurses, medical students and Ph.D students from our institute who volunteered for nasal and pharyngeal sample collection and completed a questionnaire addressing demographics, chronic medical conditions, substance abuse, vaccination status, respiratory symptoms and signs at enrolment. This assessment was advertised only in the year 2018 and 5 days after the homeless people recruitment. Controls were selected in order to avoid marked differences in terms of age, gender or origin with the homeless group. The Méditerranée Infection Institute is at the front of the fight against health care-associated infections. The building was conceived with a specifically designed setting aiming at reducing at-risk contacts with patients.13 Staffs in clinical wards are controlled through automated continuous monitoring systems for the traceability of care and good practice reminders have been developed to an anthropological approach. In this context, the control group is unlikely to carry more resistance genes because of working at the institute.14
DNA extraction and pool
A pair of swabs (nasal and pharyngeal) were collected from each participant, transferred to Sigma-Virocult® medium and were subsequently processed for molecular analysis. The automated DNA extraction was performed on 200 µL of each swab using a BioRobot®EZ1 Advanced XL instrument (QIAGEN, Hilden, Germany) and DNeasy® Blood & Tissue according to the manufacturer’s instructions. DNA pooling was performed as previously described.15 The DNA extraction quality of each pool was assessed by RT-PCR targeting internal control TISS phage that was added to each extraction.16
Real-time PCR
All quantitative real-time PCR (qPCR) reactions were performed using a C1000 Touch™ Thermal Cycle (Bio-Rad, USA) with the ready-to-use reaction mix ROX qPCR Master according to the manufacturer’s recommendations. Negative control (single PCR mix and sterile H2O) and positive control template (Plasmid DNA extracted from a colony of Escherichia coli or K. pneumoniae cultured) were included in each qPCR experimental run. Results were considered positive accepted when the cycle threshold value of real-time PCR was ≤35. Individual retesting of each specimen was carried out from positive pools. Eventually, individuals having at least a nasal or a pharyngeal positive sample were considered positive cases.
The qPCR amplification was used to confirm the presence of 1) most common β-lactamase encoding genes including blaTEM, blaSHV, ESBL genes (blaCTX-A and blaCTX-B [blaCTX-M clusters A and B]) and carbapenemase-encoding genes (blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-58, blaNDM, blaVIM, blaKPC), and 2) colistin-resistance genes (mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5) by using primers as described and by using specific primers designed in our laboratory (Table 1).17,18
 | Table 1 Sequences of primers and probes used for real-time PCRs and conventional PCRs in this study |
Conventional PCR and sequencing
To better characterize these genes, only positive qPCR results were simultaneously tested by standard PCR. The purified PCR products were sequenced using specific primers and the BigDye Terminator® version 1.1 cycle sequencing ready reaction mix (Applied Biosystems, Foster City, CA, USA). All primers used in this study have previously been described (Table 1).17,18 Sequencing was performed on Applied Biosystems 3130 platform (ABI PRISM, PE Applied Biosystems, USA). Obtained sequences were edited and assembled using Chromas Pro 1.77 (Technelysium Pty Ltd, Australia) and were then aligned with reference genes from the ARG-ANNOT by Mega 7.0 software (
Statistical analysis
Collected data were statistically treated using Microsoft Excel 2016 and STATA (version 11.1). Missing data and unidentified samples were not analyzed. Statistical differences in baseline characteristics were evaluated by Pearson’s chi-square or Fisher’s exact tests as categorical variables. Means of quantitative data were compared using Student’s t-test. A two-tailed p-value <0.05 was considered as statistically significant. We first compared the prevalence of at least one resistance gene between the homeless group and the control group (in the year 2018). In addition, two logistic regression models were applied in order 1) to compare the distribution of potential risk factors among homeless and control groups (in the year 2018); 2) to identify factors associated with resistance gene carriage in the homeless group (in the 2014–2018 period). In the first model, univariate analysis was used to examine unadjusted distributions of multiple factors (demographic, chronic medical condition), respiratory symptoms or physical finding between. A p-value <0.05 was considered statistically significant. Variables with p-values of <0.2 from the univariate analysis were included in the multivariable multinomial regression, which was then created by step-wise regression. Using the same methods, in the second model, the same set of potential risk factors together with special characteristic for homeless (type of housing, duration of homelessness) were considered for association with resistance genes. Variation over time was adequate (assessed statistically as the proportion of variance explained by year and considered adequately fitted if the coefficient of determination [R2 statistic] was >50%) by using Microsoft Excel 2016.
Results
A total of 724 homeless people was included in the study, of whom 715 (98.8%) had a nasal and/or pharyngeal swab. In the comparison group, 54 individuals were recruited and all provided both nasal and pharyngeal swabs. A total of 1,530 respiratory samples was collected. The DNA was grouped into 87 pools (15 pools of 10, 12 pools of 15 and 60 pools of 20).
Participant characteristics
The homeless individuals were predominantly male (98.2%) with a median age of 43 years (ranging 18–84 years). Fifty-five percent of them were recruited from shelter A and 45% from shelter B. Participants originated from 49 countries with a majority of African migrants (70%). About 54% of the migrant population arrived in France more than a year earlier, with a mean (SD) length of stay in France of 10 years. Overall, the average duration of homelessness was about 3 years. A proportion of 9.6% of homeless people reported chronic respiratory disease and 60.3% smoking tobacco. About 40% were suffering from at least one respiratory symptom or sign at the time of sampling, with a 27.4% cough prevalence. Vaccination rate against influenza was less than 15%. Other socio-demographic characteristics, substance abuse, chronic diseases and clinical features of participants are shown in Table 2.
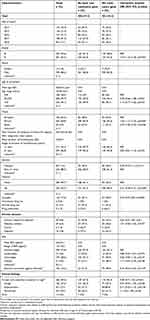 | Table 2 Characteristics of homeless participants: Demographics, chronic medical conditions and respiratory for resistance gene carriage (N=724 individuals) |
Controls were significantly more likely to be vaccinated against influenza compared to homeless people enrolled in 2018. Homeless people were significantly more likely to report chronic diseases and tobacco smoking and to present with respiratory symptoms at sampling time, compared to controls (Table 3). The differences in the distribution of several factors between the two groups remained significant in multivariate multinomial regression, such as tobacco smoking, respiratory clinical signs and vaccination against influenza (Table 4).
 | Table 3 Comparison of homeless people and controls (year 2018) |
 | Table 4 Multivariate analysis of distribution of demographics, chronic medical conditions, clinical finding between the two groups (homeless people versus controls) |
Screening of β-lactamase encoding genes (Table 5 and Figure 1)
blaTEM
Among the pools tested by qPCR screening, 82.8% (72/87) were positive for blaTEM. By individual retesting of each specimen for positive pools, we found a prevalence of 54.7% (396/710) in the homeless group over a 5-year period. Interestingly, in the control group, the prevalence was of 72.2% in 2018 (39/54), which was significantly higher than in the homeless group recruited in 2018 (52%, 51 of 98, p<0.001). Of note, using sequencing reaction targeting blaTEM in the 72 pools testing positive for blaTEM, we succeeded in amplifying 69 sequences and obtained 5 sequence types from the homeless group and 2 from the comparison group, showing 99–100% nucleotide identity to blaTEM type/reference genes in ARG-ANNOT site (Figure 1A).
 | Table 5 Prevalence of antibiotic resistance genes in nasal/pharyngeal samples in homeless population in the period 2014–2018 (N=715 individuals) |
blaSHV
About 18.4% (16/87) of the pools were positive for blaSHV-qPCR. In the homeless group, blaSHV prevalence was 3.8% (27/708). There was no significant difference of prevalence between the two groups in 2018 (7/98 in homeless people versus 4/54 in controls, p=0.92).
For the genotypic identification of blaSHV, we succeeded in amplifying 25 sequences (of 27, 92.6%) for homeless population and 4 (of 4, 100%) for the comparison group. The sequence results show the diversity of blaSHV in the homeless group (comprising 11 sequence types) and in the control group (comprising 3 sequence types) (Figure 1B).
blaCTX-M-A and blaCTX-M-B
None of the DNA pools tested positive for blaCTX-M-A and blaCTX-M-B.
Carbapenemase-encoding genes
Only 1 homeless (0.14%) was positive for blaOXA-23 by both qPCR and sequencing in nasal swab sampled in 2014 (Figure 1C). None of the DNA pools tested positive for blaOXA-58, blaOXA-48, blaOXA-24, blaNDM, blaVIM.
Screening of colistin-resistance genes
None of the DNA pools tested positive for mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 genes.
Overall, by qPCR, 57.5% (408/710) samples were positive for at least one resistance gene (blaSHV or/and blaTEM or/and blaOXA-23) in the homeless group; the proportion of blaSHV (or blaTEM) genes did not significantly vary over time (Figure 2). A higher prevalence of at least one resistance gene was observed in the comparison group (40 of 54, 74.1%) compared to the homeless group (56 of 98, 57.1%, p=0.038) (Figure 3).
 | Figure 2 Prevalence of blaTEM and blaSHV according to the year of study (homeless group). |
 | Figure 3 Percentage of individuals harboring antibiotic resistance genes (Comparison between two groups; homeless group: N=98 individuals; controls: N=54 individuals). |
Factor associated with resistance genes prevalence in the homeless population: multivariate model
Resistance genes prevalence was significantly higher in participants housed in shelter A (compared to shelter B) and in those born in African or Asian countries compared to European countries. Individuals smoking tobacco and cannabis were less likely to present resistance genes than others. In multivariate analysis, only being housed in shelter A (OR=1.59 [1.14–2.2], p=0.006) and smoking tobacco (OR=0.55 [0.39–0.78], p=0.001) remained associated with resistance gene carriage (Table 6).
 | Table 6 Multivariate analysis of demographics, chronic medical conditions, clinical findings for resistance gene carriage in homeless group in the period 2014–2018 (N=715 individuals) |
Discussion
A report from the Public Assistance – Hospitals of Marseille estimated that there are approximately 1,500 homeless individuals, including 800 sleeping on city streets, park benches, and subway trains and approximately 600 residing temporarily at the 2 main municipal shelters, with a high turnover.20 This is the first retrospective study aiming to assess directly resistance gene carriage rates in nasal/pharyngeal samples among sheltered homeless and their potential risk factors. As mentioned in several studies, tobacco smoking and suffering of respiratory symptoms at inclusion were strongly associated with homelessness status.21–23 By contrast, the prevalence of seasonal vaccination against influenza was lower in the homeless group compared to controls; in fact, the seasonal vaccination coverage in homeless people ≥65 years was only 32.9% compared to 48.5–50.8% in the overall general French population ≥65 years in the same period.24
Homeless people from shelter A were more likely to carry resistance genes compared to those from shelter B. A sub-population of homeless people with a high level of precariousness is housed in a special sector of shelter A, which may partially explain our results. Unfortunately, the fact of being housed in the special unit was not documented on a regular basis in our surveys. We also found a negative association between tobacco smoking and resistance gene carriage. Tobacco has been shown to have impact on the nasopharyngeal flora of smokers,25 which may possibly account for the lower prevalence of resistance gene in homeless smokers, although further studies are needed before conclusions can be drawn.
Overall, a lower proportion of β-lactamase encoding gene carriage was observed among the homeless individuals compared to controls. A possible explanation is the limited access to health care and subsequently reduced exposure to antibiotics in homeless people, since, in France, antibiotics are not available for sale without prescription. Further longitudinal studies are needed to better assess the antibiotic use in this population and to challenge this hypothesis.
To date, more than 400 members of blaTEM and blaSHV have been described.26 It is proven that blaTEM-1 is one of the major plasmids associated with H. influenzae which is a major causative bacterium of community-acquired respiratory tract infections.27 In a previous work, a high prevalence of Haemophilus influenzae (59%) was shown in this population during the period 2015–2017.28 In this study, similarly high genetic diversity among blaTEM-encoding strains was observed; each blaTEM has one (blaTEM-2, blaTEM-29, blaTEM-55, blaTEM-59, blaTEM-96, blaTEM-171) amino acid substitution when compared to blaTEM-1.26 For blaSHV-1, a non-ESBL variant was first identified as plasmid-encoded in E. coli from Switzerland and subsequently described worldwide in isolates in different epidemiological settings, both in humans and animals.29,30 The genetic diversity of blaSHV-encoding gene in our study did not correlate with the origin of migrants (data not shown).
We reported a very low prevalence of carbapenemase-encoding genes (only blaOXA-23 with 1 of 708 individuals, 0.14%). In surveys conducted among healthy French pilgrims before departure to the Hajj pilgrimage, the prevalence of carbapenemase-encoding was also low (1% blaOXA-48) when detected by the same molecular method.18
Since the first description of mobilized colistin gene mcr-1 in a plasmid carried by E. coli isolated in China in April 2011,31 the dissemination of the transposon has been reported in numerous countries across five continents.32 Few data are available on the prevalence of mcr-genes other than mcr-1 in the human sample. In France, to our knowledge, mcr-genes others than mcr-1 have rarely been reported,33–37 and no data is available concerning the presence of mcr-2 to mcr-5 in human samples.
Our study was based on non-culture techniques for screening antimicrobial resistance. Because many resistance genes are located on plasmids, resistance screening by direct qPCR provides a quick and simple method of screening products from genetic manipulations and a rapid estimation of antimicrobial resistance. Furthermore, pooling DNA reduces the cost of real-time PCR and yields a high specificity and a high sensitivity.16
Our work has several limitations. Homeless population was not randomly selected so that those harboring respiratory symptoms (cough, expectoration, rhinorrhoea) might have been more prone to enroll in the survey given that a medical examination was offered. The information about antibiotic use in the past, covered only a short 2-week period, which does not allow evaluating a possible lower exposure to antibiotics in the homeless group compared to controls. The detection of resistance gene directly from specimens did not allow to identify the bacteria that housed the antibiotic resistance genes.
Conclusions
Notwithstanding these limitations, the current study evidenced an unexpected low rate of resistance gene carriage among homeless people that could be explained by limited access to health care and subsequently reduced exposure to antibiotics.
Ethics approval and informed consent
This protocol was reviewed and approved by the Marseille Institutional Review Board/Ethics Committee (Homeless population: 2010-A01406-33; Comparison group: 07-008-IFR 48). Informed consent was dated and signed by all individuals and the study was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program « Investissements d’avenir », reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI. We thank Seydina M Diene and Ngaiganam Edgarthe for their technical assistance.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wrezel O. Respiratory infections in the homeless. UWO Med J. 2009;2(78):5.
2. Faustini A, Hall AJ, Perucci CA. Factors for multidrug resistant tuberculosis in Europe: a systematic review. Thorax. 2006;61(2):158–163. doi:10.1136/thx.2005.045963
3. Ottomeyer M, Graham CD, Legg AD, et al. Prevalence of nasal colonization by methicillin-resistant Staphylococcus aureus in persons using a homeless shelter in Kansas City. Front Public Health. 2016;(4):234.
4. Leibler JH, León C, Cardoso LJP, et al. Prevalence and risk factors for MRSA nasal colonization among persons experiencing homelessness in Boston, MA. J Med Microbiol. 2017;66:1183–1188. doi:10.1099/jmm.0.000552
5. Ruhe JJ, Myers L, Mushatt D, Hasbun R. High‐level penicillin‐non-susceptible Streptococcus pneumoniae bacteremia: identification of a low‐risk subgroup. Clin Infect Dis. 2004;38:508–514. doi:10.1086/381197
6. Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4:2.
7. van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJM. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. doi:10.3389/fmicb.2011.00215
8. Antunes NT, Lamoureaux TL, Toth M, et al. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother. 2014;58(4):2119–2125. doi:10.1128/AAC.02045-12
9. Kuo SC, Lee YT, Yang SP, et al.Eradication of multidrug-resistant Acinetobacter baumannii from the respiratory tract with inhaled colistin methanesulfonate: a matched case-control study. Clin Microbiol Infect. 2012;(18):870–876. doi:10.1111/j.1469-0691.2011.03682.x
10. Hartzell JD, Kim AS, Kortepeter MG, et al. Acinetobacter pneumonia: a review. Med Gen Med. 2007;9:4.
11. Hadjadj L, Riziki T, Zhu Y, Li J, Diene S, Rolain J-M. Study of mcr-1 Gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes. 2017;8:394. doi:10.3390/genes8120394
12. Leangapichart T, Tissot-Dupont H, Raoult D, et al. Risk factors for acquisition of CTX-M genes in pilgrims during Hajj 2013 and 2014. J Antimicrob Chemother. 2017;72(9):2627–2635. doi:10.1093/jac/dkx066
13. Bataille J, Brouqui P. Building an intelligent hospital to fight contagion. Clin Infect Dis. 2017;65(Suppl1):S4–S11. doi:10.1093/cid/cix474
14. Brouqui P, Boudjema S, Soto Aladro A, et al. New approaches to prevent healthcare-associated infection. Clin Infect Dis. 2017;65(Suppl1):S50–S54. doi:10.1093/cid/cix474
15. Edouard S, Prudent E, Gautret P, Memish ZA, Raoult D, Patel R. Cost-effective pooling of DNA from nasopharyngeal swab samples for large-scale detection of bacteria by real-time PCR. J Clin Microbiol. 2015;53:1002–1004. doi:10.1128/JCM.03609-14
16. Sow D, Parola P, Sylla K, et al. Performance of real-time polymerase chain reaction assays for the detection of 20 gastrointestinal parasites in clinical samples from Senegal. Am J Trop Med Hyg. 2017;97:173–182. doi:10.4269/ajtmh.16-0781
17. Leangapichart T, Dia NM, Olaitan AO, Gautret P, Brouqui P, Rolain J-M. Acquisition of extended-spectrum β-lactamases by Escherichia coli and Klebsiella pneumoniae in gut microbiota of Pilgrims during the Hajj Pilgrimage of 2013. Antimicrob Agents Chemother. 2016;60:3222–3226. doi:10.1128/AAC.02396-15
18. Leangapichart T, Gautret P, Griffiths K, et al. Acquisition of a high diversity of bacteria during the Hajj Pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 Carbapenemase Genes. Antimicrob Agents Chemother. 2016;60(10):5942–5948. doi:10.1128/AAC.00669-16
19. Gupta SK, Padmanabhan BR, Diene SM, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi:10.1128/AAC.02045-12
20.
21. Collins SE, Orfaly VE, Wu T, et al. Content analysis of homeless smokers’ perspectives on established and alternative smoking interventions. Int J Drug Policy. 2018;51:10–17. doi:10.1016/j.drugpo.2017.09.007
22. van Laere I, de Wit M, Klazinga N. Shelter-based convalescence for homeless adults in Amsterdam: a descriptive study. BMC Health Serv Res. 2009;18:208. doi:10.1186/1472-6963-9-208
23. De Maio G, Van Den Bergh R, Garelli S, et al. Reaching out to the forgotten: providing access to medical care for the homeless in Italy. Int Health. 2014;6:93–98. doi:10.1093/inthealth/ihu002
24.
25. Brook I, Gober AE. Recovery of potential pathogens and interfering bacteria in the nasopharynx of smokers and non-smokers. Chest. 2005;127:2072–2075. doi:10.1378/chest.127.2.630
26.
27. Tristram S, Jacobs MR, Appelbaum PC. Antimicrobial resistance in Haemophilus influenzae. Clin Microbiol Rev. 2007;20:368–389. doi:10.1128/CMR.00040-06
28. Ly TDA, Edouard S, Badiaga S, et al. Epidemiology of respiratory pathogen carriage in the homeless population within two shelters in Marseille, France, 2015–2017: cross sectional 1-day surveys. Clin Microbiol Infect. 2019;25(2):e1–249.e6. doi:10.1016/j.cmi.2018.04.032
29. Liakopoulos A, Mevius D, Ceccarelli D. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front Microbiol. 2016;7:1374. doi:10.3389/fmicb.2016.01374
30. Heritage J, M’Zali FH, Gascoyne-Binzi D, Hawkey PM. Evolution and spread of SHV extended-spectrum β-lactamases in Gram-negative bacteria. J Antimicrob Chemother. 1999;44:309–318. doi:10.1093/jac/44.3.309
31. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi:10.1016/S1473-3099(16)30197-9
32. Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179. doi:10.1038/s41467-018-03205-z
33. Rolain J-M, Kempf M, Leangapichart T, et al. Plasmid-mediated mcr-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob Agents Chemother. 2016;60:6994–6995. doi:10.1128/AAC.00960-16
34. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi:10.1128/CMR.00064-16
35. Beyrouthy R, Robin F, Lessene A, et al. MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob Agents Chemother. 2017;61(8):e02540–16. doi:10.1128/AAC.02540-16
36. Baron S, Bardet L, Dubourg G, et al. MCR-1 plasmid-mediated colistin resistance gene detection in an Enterobacter cloacae clinical isolate in France. J Glob Antimicrob Resist. 2017;10:35–36. doi:10.1016/j.jgar.2017.05.004
37. Birgy A, Madhi F, Hogan J, et al. CTX-M-55, MCR-1 and FosA-producing multidrug-resistant Escherichia coli infection in a child in France. Antimicrob Agents Chemother. 2018;62(4):e00127–18. doi:10.1128/AAC.00127-18
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

