Back to Journals » OncoTargets and Therapy » Volume 12
Low microRNA-622 expression predicts poor prognosis and is associated with ZEB2 in glioma
Authors Song Q, Pang H, Qi L, Liang C, Wang T, Wang W, Li R
Received 3 June 2019
Accepted for publication 15 August 2019
Published 10 September 2019 Volume 2019:12 Pages 7387—7397
DOI https://doi.org/10.2147/OTT.S218161
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Qian Song,1 Honggang Pang,2 Lei Qi,1 Chen Liang,1 Tuo Wang,1 Wei Wang,1 Ruichun Li1
1Department of Neurosurgery, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi 710061, People’s Republic of China; 2Department of Peripheral Vascular Disease, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi 710061, People’s Republic of China
Correspondence: Ruichun Li
Department of Neurosurgery, The First Affiliated Hospital of Xi’an Jiaotong University, No. 277 Yanta West Road, Xi’an, Shaanxi 710061, People’s Republic of China
Email [email protected]
Background: MicroRNAs have been recently reported to play an important role in tumorigenesis and progression in several forms of tumors. Previous studies have shown that microRNA-622 (miR-622) was associated with glioma proliferation and invasion. However, the clinical significance of miR-622 in glioma has not been elucidated. The aim of our study was to investigate the clinical values of miR-622, as well as investigate the potential molecular mechanisms in glioma.
Materials and methods: qRT-PCR and Western blot analysis were used to analyze the expression of miR-622 and ZEB2, respectively. Kaplan–Meier analysis and Cox’s proportional hazards model were used in survival analysis. MTT assay, wound healing assay, transwell assay and flow cytometry analysis were carried out to detect the impact of miR-622 on glioma cell proliferation, migration, invasion and apoptosis.
Results: Our result indicated that miR-622 expression was greatly decreased in glioma tissues and cell lines and the downregulation of miR-622 was significantly associated with the advanced pathological grade and low Karnofsky performance score of glioma. In addition, Kaplan–Meier curves with log-rank analysis revealed a close correlation between downregulation of miR-622 expression and low overall survival rate in glioma patients. Furthermore, Cox regression analysis demonstrated that downregulated miR-622 could be considered as an independent poor prognostic indicator in glioma patients. Finally, our findings demonstrated that miR-622 overexpression remarkably suppressed glioma cell proliferation, migration and invasion, while facilitated apoptosis by suppressing ZEB2 in vitro.
Conclusion: Our study suggested that miR-622 may be identified as a valuable prognostic biomarker and a promising therapeutic target for glioma patients.
Keywords: glioma, microRNA-622, prognosis, overall survival, ZEB2
Introduction
Glioma is the most common and lethal type of human primary brain tumor in adults, accounting for approximately 80% of primary malignancies of the brain, and it contributes to high mortality and morbidity while low survival rate.1,2 Based on the classification of WHO, glioma is divided into four histopathologic grades (WHO I, pilocytic astrocytoma; WHO II, diffuse astrocytoma; WHO III, anaplastic astrocytoma; and WHO IV, glioblastoma), corresponding to the increasing level of malignancy.3 Unfortunately, despite the current treatment joints multiple therapies, including surgery, radiotherapy and chemotherapy, patients, especially those who are diagnosed with glioblastoma, have an extremely poor prognosis with the median survival time of only 12–15 months and the 5-year survival rate <10%, due to its inherently progressive overgrowth, expansively migration and highly invasive nature.4,5 In recent years, some of the molecular and biological risk factors for glioma had been confirmed.6–9 Nevertheless, only few of such signatures could be identified to predict the clinical prognosis for glioma patients and finally developed new therapeutic targets. Therefore, to better recognize the pathogenic mechanism of glioma, and discover new prognostic and therapeutic value of targets are crucial for glioma patients.
MicroRNAs (miRNAs) are a set of endogenous, small non-coding RNA (19–22 nucleotides), regulating protein-coding genes expression by binding to the 3′-untranslated region (3′-UTR) of target mRNA, thus resulting in cleavage of such mRNA or inhibition of its translation.10,11 Emerging evidence strongly suggested that miRNAs are frequently dysregulated in tumors, and the dysregulation of miRNAs might involve in a variety of biological and pathological activities, such as tumor cells proliferation, migration, invasion, apoptosis, cell cycle regulation and angiogenesis.12–15 These findings mean that miRNAs might function as oncogenes and/or tumor suppressors, and might be closely associated with cancer development or suppression.16–19 Therefore, miRNAs seem to be a potential tumor therapeutic target. Notably, increasing studies have demonstrated that the aberrant expression of some miRNAs plays an important role in human glioma, which may predict prognosis for glioma patients. For example, it is reported that the decreased expression of miR-200b,20 miR-34a,21 miR-20322 and miR-145,23 and the increased expression of miR-130b,24 miR-21,25 miR-10b26 and miR-65027 are associated with poorer clinical outcome in glioma patients. In the present study, we focus on miR-622, a novel member of miRNAs, which has been identified as a potent tumor suppressor by targeting critical cancer-related pathways. Accumulating evidence has showed a decreased expression of miR-622 in hepatocellular carcinoma,28 esophageal squamous cell carcinoma,29 ovarian cancer,30 gastric cancer31 and colorectal cancer.32 More recently, Zhang et al found that miR-622 suppresses glioma cells proliferation, invasion and migration by directly targeting activating transcription factor 2.33 Of note, similar outcome was observed in another study, as reported that upregulated miR-622 expression level inhibited cell proliferation, motility and invasion in glioblastoma.34 However, to the best of our knowledge, there is no study to report the clinical significance of miR-622 associated with clinicopathologic characteristics and prognosis in glioma yet.
To address such issue, in the present study, the expression level of miR-622 in brain tissues and cells was tested by quantitative real-time reverse transcriptive-PCR (qRT-PCR). In addition, the association of miR-622 with clinicopathological characteristics and patient prognosis was analyzed. We finally investigated the effect of miR-622 on glioma cell proliferation, migration, invasion and apoptosis, and explored the potential molecular mechanisms. Our findings suggest that miR-622 may serve as a potential and valuable prognostic predictor, and a novel therapeutic target for glioma.
Materials and methods
Patients and tissue samples
The clinical specimens were collected from 108 glioma patients who underwent surgical resection, and 36 non-neoplastic patients who suffered from trauma surgery between January 2009 and November 2012 at the Department of Neurosurgery, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi Province, People's Republic of China. None of these patients had ever received chemotherapy or radiotherapy prior to surgery, and no bias against the selection for the tissue specimens was introduced in this study. Then, all fresh brain samples were immediately frozen in liquid nitrogen at −80°C for RNA extraction. Glioma patients were confirmed by histological examination, and classified according to WHO classification system, including 25 patients with pilocytic astrocytoma (WHO grade I), 21 patients with diffuse astrocytoma (WHO grade II), 32 patients with anaplastic glioma (WHO grade III) and 30 patients with glioblastoma (WHO grade IV). All enrolled glioma patients were followed up mainly by phone at intervals of 1 month in the initial 2 years postoperation and every 2 months thereafter, and the follow-up period lasted 60 months since the date of surgical resection. Note that the overall survival was calculated from the day of surgery to death or last follow-up. The clinicopathological features, including age, gender, Karnofsky performance score (KPS), treatment strategies, tumor diameter, tumor location, tumor number, vital status, WHO pathological grade and the corresponding treatment strategies of all glioma patients are described in detail in Table 1. The present study was approved by the Research Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University, and written informed consent was obtained from each patient before operation.
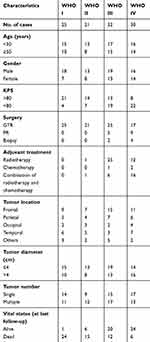 |
Table 1 The clinicopathological characteristics of 108 patients with glioma |
Glioma cell line and human astrocyte
The human glioma cell line (U251) and normal human astrocyte (NHA) were obtained from the Cell Resource Center of Shanghai Institute of Life Sciences (Shanghai, People's Republic of China) and maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated FBS (Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/mL) and streptomycin (100 µg/mL) in a humidified 5% CO2 incubator at 37°C.
Total RNA extraction and qRT-PCR analysis
Total RNA form clinical tissue specimens and cell lines were isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then, qRT-PCR was performed using an ABI 7900 Fast System (Applied Biosystems) and a TaqMan MicroRNA Assay kit (Applied Biosystems) to detect the expression of miR-622, with the small nuclear RNA U6 as a normalizing control. The primer sequences for miR-622 were as follows: 5′-ATCCCAGGGAGACAGAGATCGAGG-3′ (forward) and 5′- AAGCTTGGTGGTGGACTTTTGGTTGT-3′ (reverse). The primer sequences for U6 were 5′- CTCGCTTCGGCAGCACA-3′ (forward) and 5′- AACGCTTCACGAATTTGCGT-3′ (reverse). Further, the PCR amplification was performed as previous described. In short, the mixtures were incubated at 95ºC for 10 mins, denaturated at 95ºC for 15 s (40 cycles), annealed at 60ºC for 60 s and elongated at 72ºC for 10 mins. Each specimen was examined in triplicate and the relative expression of miR-622 in plasma was calculated and normalized using the 2−ΔΔCt method relative to U6.
Cell transfection
miR-622 mimics, a ZEB2 overexpression plasmid (ZEB2) and negative control (NC) were synthesized by RiboBio (Guangzhou, People's Republic of China). U251 cells were cultured in 6-well plates and transfected with miR-622 mimics, ZEB2 or NC with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendation.
MTT assay
U251 cells in the logarithmic phase of growth were seeded into 96-well plates. After transfection, the cell viability was tested at 24, 48, 72 and 96 hrs using the MTT assay. 50 μL of MTT solution (5 mg/mL, KeyGEN, People's Republic of China) was added into each well and the cells were incubated at 37ºC for 4 hrs. Then, we removed the medium and added dimethylsulfoxide to each well. The OD was measured at 490 nm wavelength.
Wound healing assays
For analysis of cell motility, U251 cells were cultured in 6-well plates and then incubated until 80% confluence before the conduct of wounding. Cell layers were wounded using a 200 μL tip to make a vertical wound and then washed the cells three times with PBS, and cultured with serum-free medium for 24 hrs. Cell migration to the wounded region was observed by Nikon microscopy.
Cell invasion assays
To evaluate the cell invasion ability, U251 cells (1×105) were resuspended in 150 μL serum-free medium and seeded into the upper chamber of Transwell assay inserts, while 800 μL complete medium was added to the lower chambers of each well. After incubation at 37°C for 24 hrs, cells on the upper chamber membrane were scraped off by cotton swab to remove any remaining cells. The invading cells on the lower chamber membrane were fixed with Methanol and stained with 0.1% crystal violet. Then, stained cells in five random fields were counted using a microscope. All assays were performed in triplicate.
Flow cytometry analysis for apoptosis
Transfected glioma cells were collected and washed with PBS. The cells were stained with an Annexin V-FITC/PI apoptosis detection kit (BD Pharmingen, SanDiego, CA, USA) according to the manufacturer’s instructions. The flow cytometry analysis was performed by FACS Calibur flow cytometry (BD Biosciences, USA).
Western blot analysis
U251 cells were washed with cold PBS for three times and suspended in lysis buffer. Then, the lysed samples were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% non-fat milk for 2 hrs at room temperature and incubated with ZEB2 primary antibody (Cell Signaling, Danvers, MA USA) and actin primary antibody (Abcam, Cambridge, UK) with gentle shaking at 4°C for 12 hrs. Membranes were then incubated with HRP-conjugated secondary antibody for 2 hrs in room temperature. The protein bands were visualized using ECL.
Statistical analysis
Statistical analysis was carried out using the software of SPSS version 20.0 for Windows (IBM, SPSS, Chicago, IL, USA), and the data were represented as mean±SE. One-way ANOVA test was used to evaluate the differential expression level of miR-622 and ZEB2 in glioma tissues, non-neoplastic brain tissues, U251 cells and NHA. Then, the relationship between miR-622 expression and clinicopathological features was analyzed by Pearson’s Chi-square test. Kaplan–Meier method was performed to evaluate the overall survival rate, and the difference in overall survival was estimated using the log-rank test. The Cox proportional hazards regression model was applied for univariate and multivariate analysis to calculate the HR and the 95% CI, and assess the effects of miR-622 and clinicopathological characteristics on overall survival rate. Differences were considered statistically significant when p<0.05.
Results
miR-622 expression level was downregulated in human glioma tissues and cell line
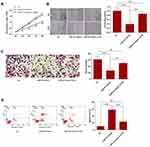
To determine the relative expression level of miR-622 between glioma tissues and normal brain tissues, the expression of miR-622 was evaluated in 108 glioma and 36 non-neoplastic brain tissues by qRT-PCR. The result showed that miR-622 expression level was significantly decreased when compared with that in non-neoplastic tissues (glioma, 3.99±0.12 vs non-neoplastic tissues, 6.33±0.20, p<0.001; Figure 1A). In addition, we also found that miR-622 expression level showed a distinctly downward tendency along with the increasing malignancy degree of glioma. Of note, as shown in Figure 1B, there was no statistical difference between grade I and grade II (grade I, 4.78±0.18 vs grade II, 4.39±0.28, p=0.24), and between grade III and grade IV (grade III, 3.29±0.16 vs grade IV, 2.94±0.17, p=0.14), respectively. It should also be noted that the expression of miR-622 in grade III was significantly lower than that in grade II (grade II, 4.39±0.28 vs grade III, 3.29±0.16, p<0.001). Furthermore, the expression level of miR-622 in U251 cells and NHA was examined. In line with the tendency in tissues, we found a similarly robust decrease of miR-622 expression in tumor cells compared to that in NHA (Figure 1C).
Relationship between miR-622 expression level and clinicopathological characteristics in glioma patients
We then explore the association of miR-622 expression with various clinicopathological features, including age, gender, WHO pathological grade, KPS, tumor diameter, tumor location and tumor number in 108 glioma patients. Fifty-five patients were assigned to the low expression group (mean expression value 3.9), with miR-622 expression level less than the median expression level (mean expression value 2.9). In contrast, the other 53 patients were assigned to the high expression group (mean expression value 5.1), with expression above the median expression level. As shown in Table 2, there was a close correlation between low miR-622 expression level and the advanced WHO pathological grade of glioma (p=0.002) and low KPS (p=0.012). However, miR-622 expression level was not significantly associated with age, gender, tumor diameter, tumor location or tumor number (p=0.254, 0.583, 0.234, 0.618 and 0.339, respectively).
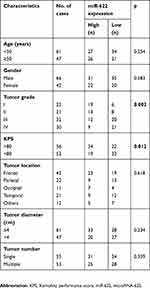 |
Table 2 Relationship between miR-622 expression level and clinicopathological characteristics in glioma patients |
Low expression level of miR-622 was significantly associated with decreased overall survival in patients with glioma
In order to evaluate the prognostic value of miR-622 for glioma, Kaplan–Meier curves with log-rank analysis were performed to investigate the relationship between miR-622 expression levels and overall survival in 108 glioma patients. Kaplan–Meier survival curves suggested that low level of miR-622 expression was significantly associated with decreased overall survival of glioma patients (p=0.002; Figure 2). Furthermore, univariate analysis using a Cox proportional hazard regression model was carried out to assess whether miR-622 expression and other clinicopathological characteristics are prognostic biomarkers for glioma patients. The data showed that low expression level of miR-622 (HR=2.701, 95% CI: 1.636–5.372, p=0.002), the advanced WHO pathological grade of glioma (HR=2.171, 95% CI: 1.235–3.811, p=0.021) and low KPS (HR=1.967, 95% CI: 1.147–3.438, p=0.039) were the prime variables for glioma prognosis (Table 3). After adjusting for clinicopathological characteristics, low expression level of miR-622 (HR=3.525, 95% CI: 2.154–6.778, p<0.001), the advanced WHO pathological grade of glioma (HR=2.682, 95% CI: 1.513–5.154, p=0.005) and low KPS (HR=2.441, 95% CI: 1.473–4.686, p=0.013) remained significantly correlated with poor prognosis of glioma patients (Table 3). Taken together, our findings demonstrated that low expression of miR-622 remained significantly correlated with worse clinical outcomes and may function as a novel and independent indicator of poor prognosis of glioma patients.
 |
Table 3 Univariate and multivariate analyses of different clinicopathological variables and miR-622 expression in patients with glioma |
Overexpression of miR-622 significantly inhibited cell proliferation, migration and invasion of glioma cells
To further investigate the role of miR-622 in glioma, experiment in vitro was performed. U251 cells were transfected with miR-622 mimics or NC, respectively. Then, MTT assay was conducted to assess the effects of miR-622 on cell proliferation at 24, 48, 72 and 96 hrs after transfection. The result showed that overexpression of miR-622 significantly inhibited cell proliferation (Figure 3A). Furthermore, the effects of miR-622 on cell motility of U251 cells were detected by wound healing assay. We found that the migration in U251 cells transfected with miR-622 mimics was obviously impeded compared to NC group (Figure 3B). Moreover, the number of invaded cells was remarkably attenuated in the miR-622 mimics group compared with NC group (Figure 3C). Additionally, flow cytometric analysis showed that the apoptosis rate of U251 cells was significantly increased in the miR-622 mimics group compared with NC group (Figure 3D). Taken together, these results indicated that miR-622 might serve as a tumor suppressor in regulating glioma biological process.
miR-622 functions as a tumor suppressor by suppressing ZEB2 in glioma
We then explored the underlying molecular mechanisms of the tumor-suppressive effect of miR-622 on glioma. Here, we first measured the expression of ZEB2 in our samples. The results indicated that ZEB2 expression was significantly upregulated in glioma tissues and cell line compared to that in non-neoplastic brain tissues or NHA, respectively (Figure 4A and B). Then, U251 cells were transfected with NC or miR-622 mimics, respectively. We found that overexpression of miR-622 could significantly reduce both ZEB2 mRNA and protein expression level (Figure 4C and D). Further, MTT assay, wound healing assay, transwell assay and flow cytometry analysis were performed to evaluate the impact of those interventions on glioma cell proliferation, migration, invasion and apoptosis. Our findings revealed that miR-622 overexpression dramatically suppressed the proliferation, migration and invasion, while facilitated apoptosis of glioma cells. Whereas the tumor-suppressive effects of miR-622 in miR-622 mimics group were reversed after co-transfection with ZEB2, indicating that ZEB2 is the downstream target gene of miR-622, and miR-622 inhibited malignant progression of glioma by suppressing ZEB2 in vitro (Figure 3A–D).
Discussion
Glioma, especially glioblastomas, is the most devastating malignant tumor in central nervous system, characterized by rapid infiltrative growth, and diffuse metastasis. As a result, glioma patients have a poor overall survival rate and a high mortality rate, despite the great advances in tumor diagnosis and treatment.35 Until now, several clinicopathologic factors, including glioma grades and KPS, are used for determining glioma patient prognosis in clinical practice. However, accurately predicting prognosis for patients with malignant gliomas remains poor. Therefore, it is of great significance to identify more potential and reliable prognostic value of biomarkers for glioma patients. Recently, accumulating evidence has demonstrated that the aberrant expression of various miRNAs is involved in the process of initiation and development in multiple forms of tumors, advocating the dysregulation of miRNAs, downregulated or overregulated, as novel prognostic biomarkers and therapeutic targets.36
In this study, we investigated the prognostic role of miR-622, one of the cancer-related miRNAs, in human glioma patients, and evaluated if the biomarker could predict disease relapse and patient outcomes. Previous studies have shown that downregulated miR-622 is closely related to malignant tumor aggressiveness and metastasis, including glioma. Cheng et al found that upregulated miR-622 repressed tumor metastasis by suppressing hypoxia-inducible factor-1alpha in ERK-responsive lung cancer.37 However, whether miR-622 is associated with clinical outcome in glioma patients is still unknown. Therefore, in the present study, we explored the correlation of miR-622 expression with glioma prognosis. Consistent with previous findings in other cancers, our results showed that the expression level of miR-622 was much lower in tumor tissues and cell line than that in non-neoplastic brain tissues and NHA. Of note, lower miR-622 expression level was observed in advanced pathological grades, suggesting a negative correlation with tumor malignant status. In addition, Pearson’s Chi-square test suggested low miR-622 expression level was closely associated with the advanced WHO grade and low KPS, but not with age, gender, tumor diameter, tumor location or tumor number. Subsequently, Kaplan–Meier curves with log-rank analysis suggested that low expression level of miR-622 tends to have poorer overall survival in glioma patients. Moreover, univariate and multivariate analyses showed that miR-622 downregulation remained significantly correlated with the prognosis of glioma patients. Our data suggest that miR-622 may function as an independent prognostic biomarker of glioma. However, the exact role and underlying molecular mechanisms of miR-622 in tumorigenesis and progression of glioma have not been well characterized. Hence, further functional experiments were performed in glioma cell line.
ZEB2 is a member of the Zfh1 family of two-handed zinc finger/homeodomain proteins. Numerous studies have demonstrated that ZEB2 is a crucial regulator in promoting the initiation and development of several cancers.38,39 Kan et al showed that overexpression of miR-335 significantly inhibited cancer cell proliferation, migration and invasion by directly targeting ZEB2 in human papillary thyroid cancer.40 Moreover, Wang et al found that transcription factor E2F1 promotes epithelial-mesenchymal transition by regulating ZEB2 gene expression in small cell lung cancer.41 Until now, many genes, including E2F1, DYRK2, PDGFb, ATF2, CDK6, cyclin D2 and c-Myc, have been experimentally validated as the targets of miR-622. Of note, Qi et al revealed that downregulation of ZEB2 by siRNA significantly inhibited cell proliferation, migration, invasion and promoted cell apoptosis in glioma cells. Meanwhile, they also found that the expression of CDK4/6, Cyclin D1, Cyclin E, E2F1 and c-Myc were decreased after ZEB2 knockdown.42 Therefore, we considered if ZEB2 is the downstream target gene of miR-622. In this study, we investigated the effect of miR-622 on glioma in vitro. Our findings showed that overexpression of miR-622 significantly inhibited cell proliferation, migration and invasion, while facilitated apoptosis of glioma cells. We then explored the underlying molecular mechanisms of the tumor-suppressive effect of miR-622 on glioma. The results revealed that the expression of ZEB2 was negatively regulated by miR-622 in glioma cell. Of note, overexpression of ZEB2 could reverse the tumor-suppressive effect of miR-622 on glioma cell. Taken together, we confirmed that miR-622 inhibited malignant progression of glioma by suppressing ZEB2.
Several limitations of the present study should be noted. First, our report was a retrospective study and the sample size was relatively small. A prospective randomized study with larger sample sizes is needed to further confirm our findings. Secondly, the collection of tissue specimens was from 108 glioma patients and 36 non-neoplastic trauma patients, which led to a great difference in the number of patients in the two groups. Finally, our results suggested that decreased expression level of miR-622 was significantly associated with the malignant status and poor prognosis in glioma patients.
Conclusion
In summary, our findings, for the first time, reveal that downregulation of miR-622 is closely associated with the malignant status and poor prognosis in glioma patients. In addition, we also confirm ZEB2 is the downstream target of miR-622, and miR-622 inhibited malignant progression of glioma by suppressing ZEB2. Therefore, miR-622 may serve as a valuable prognostic marker and would be a valuable therapeutic target for glioma.
Acknowledgment
Qian Song is acknowledged as first author. We thank Professor Li, who helped us with language editing.
Ethical approval
The study was approved by the Research Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University and complied with the Declaration of Helsinki. All procedures performed in this study were in accordance with the 1964 Helsinki Declaration and its later amendments.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Visser O, Ardanaz E, Botta L, et al. Survival of adults with primary malignant brain tumours in Europe; Results of the EUROCARE-5 study. Eur J Cancer. 2015. doi:10.1016/j.ejca.2015.07.032
2. Ricard D, Idbaih A, Ducray F, et al. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. doi:10.1016/S0140-6736(11)61346-9
3. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi:10.1007/s00401-007-0243-4
4. Mcnamara S. Treatment of primary brain tumours in adults. Nurs Stand. 2012;27(14):42–47. doi:10.7748/ns2012.12.27.14.42.c9476
5. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
6. Wang Q, Zhang J, Liu Y, et al. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016;373(2):251–259. doi:10.1016/j.canlet.2016.01.039
7. Hattermann K, Fluh C, Engel D, et al. Stem cell markers in glioma progression and recurrence. Int J Oncol. 2016;49(5):1899–1910. doi:10.3892/ijo.2016.3682
8. Zheng J, Liu X, Wang P, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–1215. doi:10.1038/mt.2016.71
9. Phillips LM, Zhou X, Cogdell DE, et al. Glioma progression is mediated by an addiction to aberrant IGFBP2 expression and can be blocked using anti-IGFBP2 strategies. J Pathol. 2016;239(3):355–364. doi:10.1002/path.4734
10. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–154. doi:10.1016/S1672-0229(08)60044-3
11. Ni WJ, Leng XM. miRNA-dependent activation of mRNA translation. Microrna. 2016;5(2):83–86.
12. Zhao JJ, Yang J, Lin J, et al. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25(1):13–20. doi:10.1007/s00381-008-0701-x
13. Liu P, Zhang H, Liang X, et al. HBV preS2 promotes the expression of TAZ via miRNA-338-3p to enhance the tumorigenesis of hepatocellular carcinoma. Oncotarget. 2015;6(30):29048–29059. doi:10.18632/oncotarget.4804
14. Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review). Oncol Rep. 2015;34(6):2811–2820. doi:10.3892/or.2015.4275
15. Abe M, Bonini NM. MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol. 2013;23(1):30–36. doi:10.1016/j.tcb.2012.08.013
16. Xu H, Zhu J, Hu C, Song H, Li Y. Inhibition of microRNA-181a may suppress proliferation and invasion and promote apoptosis of cervical cancer cells through the PTEN/Akt/FOXO1 pathway. J Physiol Biochem. 2016;72(4):721–732. doi:10.1007/s13105-016-0511-7
17. Huang M, Wu L, Luo S, et al. MicroRNA-1284 inhibits proliferation and induces apoptosis in SGC-7901 human gastric cancer cells. Biotechnol Lett. 2017;39(1):33–38. doi:10.1007/s10529-016-2213-1
18. Zhang G, Jiang G, Wang C, et al. Decreased expression of microRNA-320a promotes proliferation and invasion of non-small cell lung cancer cells by increasing VDAC1 expression. Oncotarget. 2016;7(31):49470–49480. doi:10.18632/oncotarget.9943
19. Andreas E, Hoelker M, Neuhoff C, et al. MicroRNA 17-92 cluster regulates proliferation and differentiation of bovine granulosa cells by targeting PTEN and BMPR2 genes. Cell Tissue Res. 2016;366(1):219–230. doi:10.1007/s00441-016-2425-7
20. Hu S, Jiang Q, Luo D, et al. miR-200b is a key regulator of tumor progression and metabolism targeting lactate dehydrogenase A in human malignant glioma. Oncotarget. 2016;7(30):48423–48431. doi:10.18632/oncotarget.10301
21. Guessous F, Zhang Y, Kofman A, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9(6):1031–1036. doi:10.4161/cc.9.6.10987
22. Dontula R, Dinasarapu A, Chetty C, et al. MicroRNA 203 modulates glioma cell migration via Robo1/ERK/MMP-9 signaling. Genes Cancer. 2013;4(7–8):285–296. doi:10.1177/1947601913500141
23. Lee HK, Bier A, Cazacu S, et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8(2):e54652. doi:10.1371/journal.pone.0054652
24. Gu JJ, Zhang JH, Chen HJ, Wang -S-S. MicroRNA-130b promotes cell proliferation and invasion by inhibiting peroxisome proliferator-activated receptor-gamma in human glioma cells. Int J Mol Med. 2016;37(6):1587–1593. doi:10.3892/ijmm.2016.2580
25. Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. doi:10.1128/MCB.00479-08
26. Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125(6):1407–1413. doi:10.1002/ijc.24522
27. Sun B, Pu B, Chu D, Chu X, Li W, Wei D. MicroRNA-650 expression in glioma is associated with prognosis of patients. J Neurooncol. 2013;115(3):375–380. doi:10.1007/s11060-013-1243-y
28. Song WH, Feng XJ, Gong SJ, et al. microRNA-622 acts as a tumor suppressor in hepatocellular carcinoma. Cancer Biol Ther. 2015;16(12):1754–1763. doi:10.1080/15384047.2015.1095402
29. Song C, Lu P, Shi W, et al. miR-622 functions as a tumor suppressor and directly targets E2F1 in human esophageal squamous cell carcinoma. Biomed Pharmacother. 2016;83:843–849. doi:10.1016/j.biopha.2016.07.036
30. Choi YE, Meghani K, Brault ME, et al. Platinum and PARP inhibitor resistance due to overexpression of MicroRNA-622 in BRCA1-mutant ovarian cancer. Cell Rep. 2016;14(3):429–439. doi:10.1016/j.celrep.2015.12.046
31. Guo XB, Jing CQ, Li LP, et al. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol. 2011;17(14):1895–1902. doi:10.3748/wjg.v17.i14.1895
32. Fang Y, Sun B, Li Z, Chen Z, Xiang J. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2016;55(9):1369–1377. doi:10.1002/mc.22380
33. Zhang R, Luo H, Wang S, et al. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol. 2015;121(1):63–72. doi:10.1007/s11060-014-1607-y
34. Wang X, Xin Z, Xu Y, et al. Upregulated miRNA-622 inhibited cell proliferation, motility, and invasion via repressing Kirsten rat sarcoma in glioblastoma. Tumour Biol. 2016;37(5):5963–5970.
35. Beyer S, von Bueren AO, Klautke G, et al. A systematic review on the characteristics, treatments and outcomes of the patients with primary spinal glioblastomas or gliosarcomas reported in literature until March 2015. PLoS One. 2016;11(2):e148312.
36. Palumbo S, Miracco C, Pirtoli L, et al. Emerging roles of microRNA in modulating cell-death processes in malignant glioma. J Cell Physiol. 2014;229(3):277–286.
37. Cheng CW, Chen PM, Hsieh YH, et al. Foxo3a-mediated overexpression of microRNA-622 suppresses tumor metastasis by repressing hypoxia-inducible factor-1alpha in ERK-responsive lung cancer. Oncotarget. 2015;6(42):44222–44238.
38. Nagaishi M, Nakata S, Ono Y, et al. Tumoral and stromal expression of Slug, ZEB1, and ZEB2 in brain metastasis. J Clin Neurosci. 2017;46:124–128.
39. Meyer SE. From EMT to HSC to AML: ZEB2 is a cell fate switch. Blood. 2017;129(4):400–401.
40. Kan Q, Su Y, Yang H. MicroRNA-335 is downregulated in papillary thyroid cancer and suppresses cancer cell growth, migration and invasion by directly targeting ZEB2. Oncol Lett. 2017;14(6):7622–7628.
41. Wang T, Chen X, Qiao W, et al. Transcription factor E2F1 promotes EMT by regulating ZEB2 in small cell lung cancer. BMC Cancer. 2017;17(1):719.
42. Qi S, Song Y, Peng Y, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One. 2012;7(6):e38842.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.