Back to Journals » Cancer Management and Research » Volume 13
Low-Coverage Sequencing of Urine Sediment DNA for Detection of Copy Number Aberrations in Bladder Cancer
Authors Cai YX , Yang X, Lin S, Xu YW, Zhu SW, Fan DM, Zhao M, Zhang YB, Yang XX, Li X
Received 9 December 2020
Accepted for publication 15 January 2021
Published 26 February 2021 Volume 2021:13 Pages 1943—1953
DOI https://doi.org/10.2147/CMAR.S295675
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Yun-xi Cai,1,2,* Xu Yang,1,2,* Sheng Lin,3 Ya-wen Xu,4,* Shan-wen Zhu,5 Dong-mei Fan,6 Min Zhao,7 Yuan-bin Zhang,1,2 Xue-xi Yang,6 Xin Li1,2
1Shenzhen Key Laboratory of Viral Oncology, The Clinical Innovation & Research Center (CIRC), Shenzhen Hospital, Southern Medical University, Shenzhen, 518110, People’s Republic of China; 2The Third School of Clinical Medicine, Southern Medical University, Guangzhou, 510500, People’s Republic of China; 3Laboratory of Molecular Medicine, Shenzhen Health Development Research Center, Shenzhen, 518040, People’s Republic of China; 4Department of Urology, Zhujiang Hospital, Southern Medical University, Guangzhou, 510000, People’s Republic of China; 5Reproductive Medicine Center, Huizhou Central People’s Hospital, Huizhou, 516000, People’s Republic of China; 6Institute of Antibody Engineering, School of Laboratory Medical and Biotechnology, Southern Medical University, Guangzhou, 510515, People’s Republic of China; 7PANACRO (Hefei) Pharmaceutical Technology Co., Ltd., Hefei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xin Li
Shenzhen Key Laboratory of Viral Oncology, The Clinical Innovation & Research Center (CIRC), Shenzhen Hospital, Southern Medical University, 1333 Xinhu Road, Baoan District, Shenzhen, Guangdong, 518102, People’s Republic of China
Email [email protected]
Xue-xi Yang
Institute of Antibody Engineering, School of Laboratory Medicine and Biotechnology, Southern Medical University, 1838 North Guangzhou Road, Guangzhou, Guangdong, 510515, People’s Republic of China
Email [email protected]
Purpose: Chromosomal copy number aberrations (CNAs) are a hallmark of bladder cancer and a useful target for diagnostic explorations. Here we constructed a low-coverage whole-genome sequencing method for the detection of CNAs in urine sediment DNA from patients with bladder cancer.
Patients and Methods: We conducted a prospective study using urine sediment samples from 65 patients with bladder tumors, including 54 patients with bladder cancer and 11 patients with benign bladder tumors. Forty-three healthy individuals were included as normal controls. DNA was extracted from urine sediments and analyzed by low-coverage whole-genome sequencing to compare differences in CNAs among these three groups. CNAs are defined by arbitrary R values (normal range ± 2). When these values exceed ± 0.2 of normal range, gain/duplication or loss/deletion are suspected.
Results: With this method, CNAs were detected in 39 of 51 patients with bladder cancer, 2 of 10 patients with benign bladder tumors, and 8 of 39 normal controls. The lengths of DNA deletion and duplication were significantly larger in patients with bladder cancer than in patients with benign tumors or normal controls (P < 0.05). Bladder cancer duplicate CNAs mainly occurred on chromosomes 1q, 5p, 6p, 7p, 8q, and 13q, while deletions mainly occurred on 2q, 8p, 9q, 9p, and 11p. Those regions contained bladder cancer tumor-related genes, such as STK3, COX6C, SPAG1, CDKAL1, C9orf53, CDKN2A, CDKN2B, MIR31, and IFNA1. The number of CNAs detected in urine sediment DNA during the follow-up period was significantly reduced.
Conclusion: Our sequencing method is highly sensitive and can detect a minimal chromosome repeat/microdeletion change of 0.15 Mb. The use of 0.1∼ 0.3× low-coverage whole-genome sequencing can be used to detect bladder cancer CNAs in urine sediment DNA. This method provides a promising method for noninvasive diagnosis of bladder cancer, but still needs further verification in a larger sample size.
Keywords: bladder cancer, urine sediment DNA, next-generation sequence, copy number aberrations
Introduction
Bladder cancer is the ninth most common cancer worldwide, and its incidence has increased in recent years. Notably, noninvasive bladder cancer has a recurrence rate more than 30%, and the 5-year survival rate is less than 50%.1,2 According to the national Comprehensive Cancer Network bladder cancer guidelines published in 2018, the diagnosis of bladder cancer mainly relies on urinary bladder tumor resection to remove diseased tissue for pathological examination.2,3 However, this method requires invasive examination, depending on the surgeon’s judgment and experience, which may lead to missed diagnosis. Moreover, the high cost of laparoscopic surgery causes a financial burden for patients. Therefore, the development of a noninvasive, accurate preoperative examination method may reduce diagnostic dependence on invasive examinations (eg, cystoscopy) and aid in the selection of an appropriate treatment plan before surgery.
Malignant cells typically exhibit genomic instability, manifested as copy number amplification or partial/complete deletion of the entire chromosome (ie, aneuploidy or copy number aberrations [CNAs]), which may lead to cell malignant transformation through alterations related to proliferation, invasion, and metastasis.4 The bladder cancer genome possesses considerable characteristics of genomic instability, which lead to a heavy mutation burden and extensive CNAs affecting multiple chromosomes.5,6 These factors contribute to the development and progression of bladder cancer. Cytogenetic and heterozygous deletion studies of bladder cancer and cell lines have revealed many recurrent genetic aberrations. Examples include deletions of 9p and 9q in early bladder urothelial tumors;7 amplification of 1q, 3p, 6p, 8q, 10p, 17q, 20q, and deletions of 2q, 5q, 8p, 10q, 1q, 13q, 14q, 17p in T1- and T2-stage tumors; and the loss of 6q, as well as amplification of 7p and Xq, in T2–T4-stage tumors.7,8 Some of these aberrations are associated with pathological stages or outcomes of bladder cancer, which implies that they are suitable targets for use in the diagnosis of bladder cancer.9,10
Current methods for the detection of chromosomal aberrations involve the use of DNA from urine supernatant or urinary exfoliated cells. These methods include fluorescence in situ hybridization, real-time polymerase chain reaction (PCR), and microarray comparative genomic hybridization, genome-wide bisulfite sequencing,11–15 etc. Although these methods are noninvasive, they have low sensitivity, low throughput, poor efficiency, high cost, and still need multi-center validation to further develop standardized methods. Which results in limited diagnostic value, especially for early and low-grade tumors. Therefore, the investigation of noninvasive, high-sensitivity, and specific detection methods is an urgent problem in the field of bladder cancer diagnosis and treatment. With the emergence of next-generation sequencing technology, we developed a whole-genome sequencing method and analysis pipeline to identify chromosome microdeletions and microduplications. We used this approach to establish a low-coverage whole-genome sequencing-based bladder cancer detection method.
Patients and Methods
Patients and Clinical Data
Between 2016 and 2018, pretreatment midstream urine samples were collected from 65 patients with suspected malignant bladder tumors, based on the preliminary urologic and ultrasonographic examinations in Zhujiang Hospital (Guangzhou, China). The criteria for inclusion in the tumor group were initial tumor occurrence and no adjuvant treatment (eg, radiotherapy or chemotherapy before surgery). We also collected midstream morning urine samples from 43 healthy individuals for use as normal controls. Clinical data were collected for all patients and controls. All participants provided informed consent and the study protocol was approved by the Ethics Committee of Shenzhen hospital of Southern Medical University, which was undertaken according to the principles of the Helsinki Declaration of 1975 (as revised 2008).
Urine Sample Processing and DNA Extraction
Each urine sample was centrifuged at 6000 × g for 15 minutes and the resulting urine sediment was stored at −80°C. DNA extraction was performed within 2 weeks after sample collection. DNA was extracted from 2 mL of urine sediment using the Qiagen DNeasy Blood & Tissue Kit (Hilden, Germany) according to the manufacturer’s protocol.
Library Preparation, Quality Control, and Sequencing
DNA sequencing libraries were prepared using Ion Torrent library preparation kits (Ion Torrent Systems, Inc., Gilford, NH, USA) and qualified by real-time PCR using the KAPA SYBR® FAST Universal kit (Roche, Basel, Switzerland), in accordance with the manufacturer’s instructions. Library samples were then subjected to whole-genome sequencing using the Ion Torrent Proton platform (Ion Torrent Systems, Inc.), in accordance with the manufacturer’s instructions.
Sequencing Reads and Analytical Methods
The GRCh37 human genome in the NCBI database was selected as the reference sequence. Burrows Wheeler Aligner (BWA) software was used to compare sequence data from clinical samples with the reference sequence. Duplicate sequences and non-unique ratios generated by PCR were then removed. After quality control, 2 ×106 sample unique mapping reads remained. These were filtered by micro-repetition microdeletion analysis, as follows: 1) effective interval screening; 2) determination of baseline data; 3) circular binary segmentation to reduce the normalized sequence ratio, identify breakpoints, and divide different copy number intervals, and calculation of bin copy number using the following method. Finally, calculate the Zscore value of each interval. The Zscore value of each bin greater than 3 is a micro-duplication, and less than 3 is a micro-deletion.
The average value was used as the copy number of the interval. To determine whether a chromosome was abnormal, the autosomal R value was defined as the copy number minus 2, the R value of the male sex chromosome Y was the Y chromosome copy number minus 1, and the R value of the female sex chromosome X was the X chromosome copy number minus 2. A value of R > 0.2 was considered a gain/duplication, while a value <0.2 was considered a loss/deletion.
SPSS Statistics, version 20.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The Kruskal–Wallis H-test and Mann–Whitney U-test were used to compare copy number variations in urine sediment DNA among the bladder cancer, benign bladder tumor, and normal control groups. P values <0.05 were considered to indicate statistical significance.
Results
Clinicopathologic Characteristics of the Samples
Sixty-five patients with suspected bladder tumors and 43 normal controls were enrolled in this study. Bladder cancer samples were diagnosed according to surgical pathology, including bladder urothelial carcinoma and bladder adenocarcinoma. Of the 65 patients, 11 had benign bladder tumors. The clinical data for each patient are summarized in Table 1.
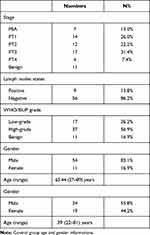 |
Table 1 Clinical Information of All Patients with Bladder Tumor |
Whole-genome low-coverage sequencing of all samples was performed on urine sediment DNA, with a median of 4.2 × 106 reads per sample, corresponding to a median reading depth of 0.1 (interquartile range, 0.08–0.16). After quality control, data were included for 100 participants, including 51 patients with bladder cancer, 10 patients with benign bladder tumors, and 39 normal controls. All sequencing statistics are shown in Table SI.
Genome-Wide Distribution and CNAs in Urine Sediments
CNAs were detected in 39 of 51 patients with bladder cancer, 2 of 10 patients with benign tumors, and 8 of 39 normal controls. Figure 1 shows the results of partial chromosome copy number variation detection for sample B1. The chromosome copy number variation data of all samples are drawn into pictures and viewed in the original and supplementary materials. The specific data for all samples are provided in Table SII and Table SIII.
 |
Figure 1 Chromosome copy number aberrations of chr6-chr9 in sample B1. |
The Kruskal–Wallis H-test was used to compare CNA-positive rates among the three groups. The bladder cancer group had a higher rate of CNA detection than the benign tumor and normal control groups (all P < 0.005), but there was no significant difference between the benign tumor and normal control groups (P = 0.55).
All data were compared with the Database of Genomic Variants (DGV) to exclude genetic copy number variation. Comparisons of the normal control group and the CNA region of the benign tumor group revealed that the N22 and N34 normal control samples showed large X chromosome repeats (>150 Mb). Although the absolute value of R did not exceed 1, the participant’s health could not be sufficiently evaluated and the genetic or clinical significance of these findings could not be determined. The CNAs of the bladder cancer group involved multiple regions, most of which were extensive deletions and amplifications of >10 Mb. These regions were also compared with the DGV, which revealed that nearly all large fragment CNA regions were absent from the DGV. These findings are provided in detail in Table SII and Table SIII.
The general area and type of copy number variation were similar among samples. The average length of deletion mutations in cancer samples was approximately 195.94 Mb, while the average length of repeated mutations was approximately 125.39 Mb. In general, the chromosome regions of bladder cancer urine sediment samples with deletions and changes were wider than those with chromosome duplication. As noted above, CNAs were detected in 2 of 10 patients with benign bladder tumors. Sample C03 exhibited 1.65-fold microduplication in the region of chr21p11.2–11.1 (length, 0.35 Mb). Additionally, CNAs were detected in 8 of 39 normal controls. Samples N22 and N34 had no more than 1-fold X chromosome duplication, the area involved was >150 Mb, and the remaining six samples had microduplications. Comparison of the lengths of repeated and missing regions in the three groups is shown in Figure 2. The length of CNA regions was much wider in the bladder cancer group than in the benign tumor group (P < 0.05) and in the normal controls (P < 0.005), but there was no significant difference in the length of CNA regions between the benign tumor and normal groups. However, the lengths of CNA regions and the positive rate of detection did not differ according to pathological or clinical stages in patients with bladder cancer.
The heatmap in Figure 3 shows the overview results of chromosomal CNAs in the 100 urine samples. All genomic regions that are overrepresented (indicative of somatic amplifications) are shown in red, whereas genomic regions that are underrepresented (indicative of somatic deletions) are shown in blue. Visual inspection showed that a large degree of chromosomal instability was present in patients with bladder cancer, while patients with benign bladder tumors had stable chromosomal patterns similar to those of normal controls.
High-Frequency CNA Regions in Urine Sediment DNA from Patients with Bladder Cancer
We grouped all CNA-positive samples into four groups according to tumor differentiation status and analyzed the frequencies of CNA regions and genetic conditions in the regions involved. Chromosome duplications were mainly identified on chromosomes 1q, 5p, 6p, 7p, 8q, and 13q; chromosome deletions were mainly identified on 2q, 8p, 9q, 9p, and a portion of the 11p arm. The frequency, region, and type of chromosomal copy number variation are shown in Figure 2.
The CNAs were classified according to the degree of bladder tumor differentiation: benign, low-grade, high-grade and control group. Low-grade potentially malignant bladder epithelial benign tumors (eg, urotheliomas and papillomas) were clustered with the benign bladder tumor group. The results are shown in Figure 4.
As above, bladder tumors were divided according to their degrees of differentiation (benign, low-grade, and high-grade). The number of samples with CNAs was higher among high-grade bladder cancers than among low-grade and benign tumors. In low-grade bladder cancer, missing copy number variation was more common, mainly in the chr9q1.12–q34.3 region, while repetitive mutations mainly occurred in the chr7q arm. High-grade bladder cancer also exhibited missing copy number variation, but the chromosomal region affected was more extensive than that of low-grade bladder cancer and benign bladder tumors. Notably, there were copy number variations in each chromosome. Deletion mutations were mainly concentrated in the p-arm of autosomal chromosome 8 and the entire segment of autosomal chromosome 9. The high-frequency regions of deletion variation were chr2q35.1–q37.3, chr8p23.3–p12, chr9q21.12–q34.3, chr11p13–p12, chr11p15.5–p15.1, chr18q12.3, and chr18q21.1–22.1. Repetitive mutations frequently occurred on the q-arm of chromosome 1, the p-arm of autosomal chromosome 5, and the p-arms of autosomal chromosomes 6 and 8. The regions with frequent repeated mutations in those chromosomes were mainly chr5p14.2–p14.1, chr6p22.3, and chr8q22.1–q22.3. These findings are shown in Table 2.
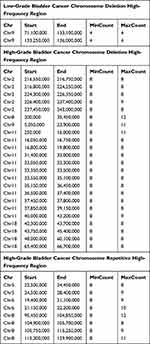 |
Table 2 Bladder Tumor Urine Sediment Chromosome Copy Number Variation High Frequency Region |
Discussion
Bladder cancer is one of the most common malignant tumors in humans, with high rates of recurrence and mortality. Studies have shown that bladder cancer has a high tumor mutation burden and considerable variations in chromosomal and gene copy numbers. Thus, copy number variation is an important target for exploration and monitoring of bladder cancer.16 In the past 30 years, urine-based pathological examination of bladder cancer has been an important auxiliary assessment in clinical practice, but this method is not particularly sensitive and has led to some missed diagnoses.11 However, exfoliated cells in urine sediment can clearly demonstrate the histopathological status of primary bladder tumor lesions. They also contain relatively complete genomic information, which is suitable for use in tumor diagnosis and testing.17 Cystoscopy remains the gold standard for diagnosis of bladder cancer, but is an invasive method. Moreover, it is prone to missed diagnosis of carcinoma in situ. Urine exfoliation cytology can avoid these problems. As noted in the Introduction, the current methods of detecting chromatin abnormalities using urinary exfoliated cells are noninvasive, but have low sensitivity, low throughput, poor efficiency, and limited diagnostic value for early and low-grade tumors.11–14,18 Following recent developments in sequencing technology, next-generation sequencing has become widely used in noninvasive prenatal testing chromosome copy number detection due to its high throughput, high sensitivity, and low cost.19–25 Therefore, we presumed that low-coverage sequencing could be used to detect chromosomal copy number variation, in combination with bladder cancer urine sediment samples to detect copy number variation in exfoliated cells from urine sediment for the diagnosis and monitoring of bladder cancer.
In this study, the low-coverage whole-genome sequencing method was used to compare the chromosomal copy number variation in urine sediment DNA from patients with bladder cancer, patients with benign bladder tumor, and normal controls. The chromosomal copy number variation in urine sediment was significantly higher in patients with bladder cancer than in patients with benign bladder tumors or normal controls. Furthermore, CNAs were considerably easier to detect in bladder cancer samples than in the other two groups. With our method, small chromosome fragments <10 Mb could be detected in urine sediment DNA from patients with bladder cancer. In these patients, large fragments of chromosomal repeat and deletion mutations were mainly concentrated on chromosomes 1q, 5p, 6p, 7p, 8q, and 13q, as well as on the arms of chromosomes 2q, 8p, 9q, 9p, and 11p. In addition, chromosomes 2q, 5q, 6q, 13q, 17q, 17p, and other regions had relatively high-frequency copy number variation. These results are largely consistent with the results of previous genome copy number variation analyses in patients with bladder cancer.7,8,26 The regions with copy number variation contain multiple genes related to the pathogenesis of cancer development, including CDKN2A, C9orf53, MIR31, FGFR3, p53, MMPs, TSP1, and COX26. These results suggest that the use of low-coverage whole-genome sequencing to detect copy number variation in urine sediment DNA is useful for noninvasive diagnosis of bladder cancer. However, data from a large number of normal controls are needed to further optimize the analysis, and large-scale prospective studies are required to confirm our findings.
Some reports have implied that the extent of copy number variation in DNA from urine or blood cfDNA is related to the tumor characteristics.17,27–29 These include reports on colon cancer, ovarian cancer, lung cancer, bladder cancer, and other tumors. With pathological progression, higher numbers of genomic CNAs and larger chromosomal regions were detected in circulating free (cf) DNA in tissue or plasma.20,21,30,31 Other studies have used droplet digital PCR, fluorescence in situ hybridization, and other techniques to detect the copy numbers of bladder cancer-related genes (eg, AURKA) in cfDNA from urine supernatant. Copy number analyses of multiple-gene panels have revealed that copy number amplification and deletion mutation rate are related to tumor progression stage.26,30 Integrated analysis has demonstrated that most bladder cancer samples, regardless of clinical stage or degree of tumor differentiation, have similar chromosomal copy number variation patterns. The higher level of tumor differentiation, the regions and types of chromosomal CNAs become more extensive. Accordingly, CNAs are more frequently detected in high-grade bladder cancer and are more likely to transition into deletion-type aberrations. These results were not related to clinical pathological stage findings. We presume that this phenomenon is related to the presence of tumor cells and other exfoliated cells in the sediment, because of repeated mutations and abnormal differentiation after the cells become cancerous. Increasing cancer cell differentiation leads to higher genomic instability and greater likelihood of CNA. However, these data do not fully support the biological model of disease progression related to molecular types of gene mutations or tumor-driven chromosomal mutations. In addition, considering that the chromosomal aberrations of urine sediment samples from bladder cancer involve a wide range, and the copy number change rate is high, but generally, genetic copy number variations are small fragments and low-fold changes, so we did not screen bladder cancer samples with DGV data during the process of biometric analysis.
During the experiment, we found that CNAs could not be detected in urine sediment DNA from some patients with high-grade or invasive bladder cancer. In those instances, we presumed that the detection was impeded by factors such as the number of exfoliated tumor cells in the urine,32 the concentration of extracted DNA, and background interference from other DNA.27 For example, noninvasive prenatal testing technology, another low-coverage sequencing method, may generate false-negative or false-positive results if maternal cfDNA causes interference during detection of fetal chromosomal aneuploidy. These outcomes are influenced by the concentration and quality of the DNA library, the depth of sequencing, the method for analysis of biological information, and the characteristics of normal controls (ie, reference samples). There are reports suggesting that by detecting the whole genome Zscore (|Z|), which calculates the degree of copy number variation of urine sediment cell DNA, can improve the specificity and sensitivity of diagnosis. It believes that the |Z| is related to tumor grade.33 This result is similar to some of ours. However, the calculation of change in genome requires a large number of urine genome samples from normal people as a negative control. Using only a small number of people or using blood genome samples as a negative control for calculation will cause data bias or even errors. Therefore, we believe that if we want to improve the low-coverage sequencing method to detect bladder cancer CNV, it is necessary to further expand the detection volume of cancer samples for stratified detection, and further improve the data of negative control samples to ensure the accuracy of the test results.
The use of cfDNA has been a popular method for noninvasive tumor detection. Many studies have suggested that cfDNA from urine or plasma supernatant in patients with bladder cancer can detect genomic characteristics similar to the primary tumor, and that these characteristics are associated with tumor stage and progression.27,34–36 Highly sensitive detection methods are needed for investigation of cfDNA in blood or urine due to its low concentrations and short peak lengths. Droplet digital PCR may be used to quantify a single mutant alleles and deep targeted sequencing may be used for assessment of multiple allele panels.25–27,37–39 Moreover, comparative genomic hybridization technology can be used to detect chromosomal aneuploidy in urinary exfoliated cells from patients with bladder cancer. Because of background interference from other DNA in urinary exfoliated cells, this method has limited sensitivity and specificity and has not been widely used.40 In contrast, whole-genome sequencing can detect a large number of biomarkers. Regarding complex non-tumor DNA background noise and low-concentration DNA, ultra-high reading depth is required to ensure accurate results.41 We attempted to use cfDNA from supernatants of blood to perform low-coverage sequencing. However, the low cfDNA concentration resulted in an extremely high rate of sequencing failures. It has been reported that the cfDNA and exosomal DNA in the urine supernatant can detect the tumor mutational burden of bladder cancer, and it has stronger sensitivity.36 There are some reports suggesting that the diagnosis efficiency can be improved by detecting the marker gene panel at the same time as detecting CNV, or combining methylation detection and other methods.36,42,43 These methods usually require higher sequencing depth and detection of tumor markers to improve diagnosis efficiency, which will increase the cost of detection, and the selection of gene panels and methylation sites requires further experimental verification. Therefore, we presume that this low-coverage sequencing method may be unsuitable for aneuploidy detection in cfDNA. To improve the success rate, both DNA concentration and sequencing depth must be increased, which will lead to greater detection cost. Additionally, collection of urine samples is more convenient in clinical practice and easier to be received by patients. Copy number changes could accumulate with age, the control group is younger than cancer patients, because the control group samples we collect are mainly from the physical examination population. So we compared the results of the control group with the DGV database (database of genomic variations), to eliminate data bias caused by human genome variation. In follow-up studies, we will expand the age range of the control group to improve the study.
In conclusion, we have shown that the use of low-coverage sequencing to detect copy number variation in urine sediment DNA is a feasible method for differential diagnosis of bladder cancer. As the cost of sequencing decreases, our noninvasive method for detection of DNA from exfoliated cells in urine sediment may be useful for diagnosing malignant tumors in the urinary system, as well as for detecting tumor recurrence and monitoring response to treatment. However, this method requires further validation with a large number of samples and additional studies to establish negative control reference ranges.
Conclusion
In summary, we found that the use of low-coverage sequencing to detect copy number variation in urine sediment DNA is a feasible method for differential diagnosis of bladder cancer. The chromosome copy number variation range and detection rate in bladder cancer is significantly different from benign and normal controls in urine sediment samples. With this method, small chromosome fragments <10 Mb could be detected in urine sediment DNA from patients with bladder cancer. High-frequency chromosomal copy number variation regions are largely consistent with the results of previous genome copy number variation analyses in patients with bladder cancer.
As the cost of sequencing decreases, our noninvasive method for detection of DNA from exfoliated cells in urine sediment may be useful for diagnosing malignant tumors in the urinary system, as well as for detecting tumor recurrence and monitoring response to treatment. However, this method requires further validation with a large number of samples and additional studies to establish negative control reference ranges.
Data Sharing Statement
Datasets supporting this paper have been deposited at the database of NCBI (https://www.ncbi.nlm.nih.gov/, project ID: PRJNA691120).
Acknowledgments
The authors thank all of the patients who participated in our study. This work was funded by grants from the National Natural Science Foundation of China (no. 81572644); the NSFC/RGC Joint Research Project (nos. N_HKU735/18 and 81861168033); the Shenzhen Key Laboratory of Viral Oncology (no. ZDSYS201707311140430); the Shenzhen Science and Technology Key Project (no. JCYJ2017041365531148); the Science and Technology Program of Guangzhou (Grant No. 201704020114). Finally, we are very grateful to the Sanming Project of Medicine in Shenzhen (no. SZSM201612023) for the contribution to this research.
Disclosure
All authors report no conflicts of interest in this work.
References
1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi:10.1016/j.eururo.2016.06.010
2. He YT, Li DJ, Liang D, et al. [Incidence and mortality of bladder cancer in China, 2014]. Zhonghua Zhong Liu Za Zhi. 2018;40(9):647–652. Chinese. doi:10.3760/cma.j.issn.0253-3766.2018.09.002
3. NCCN Guidelines Version 2.2018 Bladder Cancer.
4. Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi:10.1038/nature12965
5. Aine M, Eriksson P, Liedberg F, Sjodahl G, Hoglund M. Biological determinants of bladder cancer gene expression subtypes. Sci Rep. 2015;5(1):10957. doi:10.1038/srep10957
6. Lindgren D, Frigyesi A, Gudjonsson S, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70(9):3463–3472. doi:10.1158/0008-5472.CAN-09-4213
7. Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5(9):713–725. doi:10.1038/nrc1697
8. Choi W, Ochoa A, McConkey DJ, et al. Genetic alterations in the molecular subtypes of bladder cancer: illustration in the cancer genome atlas dataset. Eur Urol. 2017;72(3):354–365. doi:10.1016/j.eururo.2017.03.010
9. Sjodahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. doi:10.1158/1078-0432.CCR-12-0077-T
10. Cazier JB, Rao SR, McLean CM, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014;5(1):3756. doi:10.1038/ncomms4756
11. He H, Han C, Hao L, Zang G. ImmunoCyt test compared to cytology in the diagnosis of bladder cancer: a meta-analysis. Oncol Lett. 2016;12(1):83–88. doi:10.3892/ol.2016.4556
12. Patel KM, Tsui DW. The translational potential of circulating tumour DNA in oncology. Clin Biochem. 2015;48(15):957–961. doi:10.1016/j.clinbiochem.2015.04.005
13. Lodewijk I, Dueñas M, Rubio C, et al. Liquid biopsy biomarkers in bladder cancer: a current need for patient diagnosis and monitoring. Int J Mol Sci. 2018;19(9):2514. doi:10.3390/ijms19092514
14. Blick CG, Nazir SA, Mallett S, et al. Evaluation of diagnostic strategies for bladder cancer using computed tomography (CT) urography, flexible cystoscopy and voided urine cytology: results for 778 patients from a hospital haematuria clinic. BJU Int. 2012;110(1):84–94. doi:10.1111/j.1464-410X.2011.10664.x
15. Ge G, Peng D, Guan B, et al. Urothelial carcinoma detection based on copy number profiles of urinary cell-free DNA by shallow whole-genome sequencing. Clin Chem. 2020;66(1):188–198. doi:10.1373/clinchem.2019.309633
16. Tanaka K, Hirota T. Chromosomal instability: a common feature and a therapeutic target of cancer. Biochim Biophys Acta. 2016;1866(1):64–75. doi:10.1016/j.bbcan.2016.06.002
17. Togneri FS, Ward DG, Foster JM, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet. 2016;24(8):1167–1174. doi:10.1038/ejhg.2015.281
18. Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. 2015;21(21):4786–4800. doi:10.1158/1078-0432.CCR-14-1190
19. Nakabayashi M, Kawashima A, Yasuhara R, et al. Massively parallel sequencing of cell-free DNA in plasma for detecting gynaecological tumour-associated copy number alteration. Sci Rep. 2018;8(1):11205. doi:10.1038/s41598-018-29381-y
20. Cohen PA, Flowers N, Tong S, Hannan N, Pertile MD, Hui L. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med. 2016;14(1):126. doi:10.1186/s12916-016-0667-6
21. Vanderstichele A, Busschaert P, Smeets D, et al. Chromosomal instability in cell-free DNA as a highly specific biomarker for detection of ovarian cancer in women with adnexal masses. Clin Cancer Res. 2017;23(9):2223–2231. doi:10.1158/1078-0432.CCR-16-1078
22. Mersy E, Smits LJ, van Winden LA, et al. Noninvasive detection of fetal trisomy 21: systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012. Hum Reprod Update. 2013;19(4):318–329. doi:10.1093/humupd/dmt001
23. Kinnings SL, Geis JA, Almasri E, et al. Factors affecting levels of circulating cell-free fetal DNA in maternal plasma and their implications for noninvasive prenatal testing. Prenat Diagn. 2015;35(8):816–822. doi:10.1002/pd.4625
24. Noninvasive Prenatal Testing for Trisomies. 21, 18, and 13, sex chromosome aneuploidies, and microdeletions: a health technology assessment. Ont Health Technol Assess Ser. 2019;19(4):1–166.
25. Cai YH, Yao GY, Chen LJ, Gan HY, Ye CS, Yang XX. The combining effects of cell-free circulating tumor DNA of breast tumor to the noninvasive prenatal testing results: a simulating investigation. DNA Cell Biol. 2018;37(7):626–633. doi:10.1089/dna.2017.4112
26. Zhang S, Wang Y, Bondaruk J, et al. Detection of bladder cancer in urine sediments by a novel multicolor fluorescence in situ hybridization (Quartet) test. Eur Urol Focus. 2018.
27. Ward DG, Bryan RT. Liquid biopsies for bladder cancer. Transl Androl Urol. 2017;6(2):331–335. doi:10.21037/tau.2017.03.08
28. Salvi S, Casadio V. Urinary cell-free DNA: potential and applications. Methods Mol Biol. 2019;1909:201–209.
29. Christensen E, Birkenkamp-Demtroder K, Nordentoft I, et al. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur Urol. 2017;71(6):961–969. doi:10.1016/j.eururo.2016.12.016
30. Liu B, Yang L, Huang B, et al. A functional copy-number variation in MAPKAPK2 predicts risk and prognosis of lung cancer. Am J Hum Genet. 2012;91(2):384–390. doi:10.1016/j.ajhg.2012.07.003
31. Li J, Dittmar RL, Xia S, et al. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol Oncol. 2017;11(8):1099–1111. doi:10.1002/1878-0261.12077
32. Goodison S, Rosser CJ, Urquidi V. Bladder cancer detection and monitoring: assessment of urine- and blood-based marker tests. Mol Diagn Ther. 2013;17(2):71–84. doi:10.1007/s40291-013-0023-x
33. Zeng S, Ying Y, Xing N, et al. Noninvasive detection of urothelial carcinoma by cost-effective low-coverage whole-genome sequencing from urine-exfoliated cell DNA. Clin Cancer Res. 2020;26(21):5646–5654. doi:10.1158/1078-0432.CCR-20-0401
34. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. doi:10.1126/scitranslmed.3007094
35. Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. “Liquid biopsy”-ctDNA detection with great potential and challenges. Ann Transl Med. 2015;3(16):235. doi:10.3978/j.issn.2305-5839.2015.09.29
36. Lee DH, Yoon H, Park S, et al. Urinary exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci Rep. 2018;8(1):14707. doi:10.1038/s41598-018-32900-6
37. Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. 2015;48(15):948–956. doi:10.1016/j.clinbiochem.2015.03.015
38. Szarvas T, Kovalszky I, Bedi K, et al. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncol Rep. 2007;18(2):405–409.
39. Stahlberg A, Krzyzanowski PM, Jackson JB, Egyud M, Stein L, Godfrey TE. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016;44(11):e105. doi:10.1093/nar/gkw224
40. Schmitz-Drager BJ, Droller M, Lokeshwar VB, et al. Molecular markers for bladder cancer screening, early diagnosis, and surveillance: the WHO/ICUD consensus. Urol Int. 2015;94(1):1–24. doi:10.1159/000369357
41. Millholland JM, Li S, Fernandez CA, Shuber AP. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res Rep Urol. 2012;4:33–40. doi:10.2147/RRU.S32736
42. Cheng THT, Jiang P, Teoh JYC, et al. Noninvasive detection of bladder cancer by shallow-depth genome-wide bisulfite sequencing of urinary cell-free DNA for methylation and copy number profiling. Clin Chem. 2019;65(7):927–936. doi:10.1373/clinchem.2018.301341
43. Xu Z, Ge G, Guan B, et al. Noninvasive detection and localization of genitourinary cancers using urinary sediment DNA methylomes and copy number profiles. Eur Urol. 2020;77(2):288–290. doi:10.1016/j.eururo.2019.11.006
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



