Back to Journals » OncoTargets and Therapy » Volume 11
Long noncoding RNA Sox2 overlapping transcript (SOX2OT) promotes non-small-cell lung cancer migration and invasion via sponging microRNA 132 (miR-132)
Authors Zhang K, Li Y, Qu L, Ma X , Zhao H, Tang Y
Received 19 March 2018
Accepted for publication 25 May 2018
Published 30 August 2018 Volume 2018:11 Pages 5269—5278
DOI https://doi.org/10.2147/OTT.S168654
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
This paper has been retracted.
Kewei Zhang, 1,* Yang Li, 2,* Limei Qu, 3 Xiaobo Ma, 3 Hongguang Zhao, 4 Ying Tang 2
1Department of Thoracic surgery, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China; 2Department of Respiration, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China; 3Department of Pathology, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China; 4Department of Nuclear Medicine, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China
*These authors contributed equally to this work
Background: Long noncoding RNA (lncRNA) Sox2 overlapping transcript (SOX2OT) has been reported to be upregulated in various types of cancers, including non-small-cell lung cancer (NSCLC). However, the biological role and underlying mechanism of SOX2OT activity in NSCLC remain largely unknown. This study aims to investigate the function and possible molecular mechanisms of SOX2OT in NSCLC.
Materials and methods: Quantitative real-time polymerase chain reaction was used to detect SOX2OT expression, and cellular proliferation, migration, and invasion were measured using cell counting kit-8, wound healing, and Transwell invasion assays, respectively. Western blotting was used to determine protein expression. Starbase 2.0 and luciferase reporter assay were utilized to identify the molecular target of SOX2OT.
Results: Here, we discovered that SOX2OT was markedly upregulated in NSCLC tissues and cell lines. Knockdown of SOX2OT inhibited the proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT) process in NSCLC cells. Moreover, we explored the regulatory mechanism of SOX2OT and found that SOX2OT directly bound microRNA 132 (miR-132) in NSCLC cells. Importantly, miR-132 inhibition partially reversed the SOX2OT knockdown-mediated inhibitory effect on cell proliferation, migration, invasion, and EMT process. We also found that SOX2OT could regulate zinc finger E-box-binding homeobox 2 (a target of miR-132) expression, which played crucial roles in tumor cell proliferation and invasion.
Conclusion: These findings indicated that SOX2OT was a noncoding oncogene that exerted important regulatory functions in NSCLC via sponging miR-132 and might represent a novel strategy for overcoming this disease.
Keywords: non-small-cell lung cancer, SOX2OT, miR-132, ZEB2
Introduction
Non-small-cell lung cancer (NSCLC) is the most common diagnosed cancer, accounting for ~85% of all lung cancers at present.1 Although diagnostic methods and therapeutic strategies for NSCLC have been improved in the last decades, the survival rate of advanced NSCLC still remains poor mainly because of metastasis and/or postsurgical recurrence.2 Therefore, identifying the underlying molecular mechanisms involved in NSCLC progression and metastasis is critical for developing novel diagnostic biomarkers and therapeutic strategies to improve treatment outcomes in patients with NSCLC.
Long noncoding RNAs (lncRNAs) are a type of RNA transcripts <200 nucleotides in length without protein-coding capacity.3 Recently, lncRNAs have been shown to be involved in various physiological and pathological processes such as cell differentiation and apoptosis, X chromosome imprinting, and even tumorigenesis.4,5 Accumulating evidence suggests that abnormal lncRNA expression is often observed in various cancers and contributes to the initiation and development of several cancers.6,7 For NSCLC, some lncRNAs have been identified to function as oncogenes or tumor suppressors and have potential as diagnostic or prognostic markers for patients with NSCLC.8,9
Sox2 overlapping transcript (SOX2OT), a lncRNA transcribed in the same orientation as Sox2, has been shown to be upregulated and functions as oncogene in multiple types of cancers, including ovarian cancer,10 glioblastoma,11 breast cancer,12 gastric cancer,13 colorectal cancer,14 esophageal squamous cell carcinoma,15 and hepatocellular carcinoma.16 A study has shown that SOX2OT expression was upregulated in NSCLC tissues, closely associated with tumor-node-metastasis (TNM) stage, lymph node metastasis, and poor overall survival rate and that SOX2OT significantly promoted NSCLC cell proliferation by regulating cell cycle arrest at the G2/M phase.17 However, its biological function, especially in migration and invasion, and underlying regulator role in NSCLC progression remain largely unknown.
Here, we examined SOX2OT expression in NSCLC cells and its biological roles in proliferation, invasion, migration, and epithelial–mesenchymal transition (EMT) process by a series of in vitro and in vivo experiments. Moreover, we investigated the molecular mechanisms by which SOX2OT exerted its oncogene role in NSCLC.
Materials and methods
Clinical specimens
Forty-eight NSCLC tissue samples and adjacent noncancerous lung tissues were obtained from patients who underwent lung cancer surgery at the First Hospital of Jilin University (Changchun, China). Upon surgical removal of specimens, each sample was snap-frozen in liquid nitrogen and immediately frozen at −80°C until use. Written informed consents were obtained from all patients. This study was in accordance with the Declaration of Helsinki and approved by the First Hospital of Jilin University.
Cell culture and cell lines
Lung cancer cell lines (A549, H1299, NCI-H460, and HCC-827) and a human normal bronchus epithelial cell line (BEAS-2B) were obtained from the Chinese Type Culture Collection, Chinese Academy of Sciences (Shanghai, China). All cell lines were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) at 37°C with 5% CO2 and 95% air.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cells using the TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The purity of RNA was determined based on the A260/A280 ratio using ultraviolet spectrophotometry (Thermo Fisher Scientific). Total RNAs (1 μg) as the templates were transcribed into cDNA using a reverse transcription kit (Takara, Dalian, China). qRT-PCR was performed using the SYBR® Green (Takara) dye detection method on the Roche Lightcycler 96 instrument (Hoffman-La Roche Ltd., Basel, Switzerland). Primers were designed and synthetized by Takara and are listed in Table 1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as endogenous controls to detect messenger RNA (mRNA) and miRNA, respectively. The relative level was calculated using the 2−ΔΔCt method.
Cell transfections
The sequence of short-hairpin RNA (shRNA) against SOX2OT (sh-SOX2OT) and corresponding nontargeting sequences (sh-NC) were synthesized and subcloned into pGPU6/GFP/Neo vector (Gene Pharma, Shanghai, China). MicroRNA 132 (miR-132) mimic and the corresponding negative control (miR-NC) and miR-132 inhibitor and the corresponding control (anti-miR-NC) were obtained from Ribobo (Guangzhou, China). A549 and H1299 cells were transfected with the abovementioned molecular products using the Lipofectamine® 2000 reagent (Thermo Fisher Scientific) following the manufacturer’s protocol.
Cell proliferation
Cell proliferation was analyzed by using the cell counting kit-8 (CCK8) assay (Beyotime, Shanghai, China). Briefly, different groups of cells were seeded into 96-well plates at the density of 5×103 cells/well in triplicate. At indicated time (24, 48, 72, and 96 h), the cells were incubated with CCK8 reagent in the dark at 37°C for 30 min. Absorbance was measured at a wavelength of 450 nm using the Benchmark™ Plus Microplate Spectrometer (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Wound healing assay
Different groups of cells were seeded into six-well plates and grown to 100% confluence in RPMI 1640 medium. Subsequently, cell layers were wounded using the tip of a 200 μL pipette. After washing cells three times with phosphate-buffered saline (PBS), the cells were incubated in free-serum medium and cultured for 24 h. The wound closure was measured using the AxioVision software Version 4.7 (Carl Zeiss Meditec AG, Jena, Germany).
Transwell invasion assay
Cell invasion ability was determined using the Transwell insert chambers (Corning Incorporated, Corning, NY, USA) covered with Matrigel (BD Biosciences, San Jose, CA, USA). Cells were suspended in serum-free medium and seeded into the upper chamber at a density of 2.0×104 cells/well, while RPMI 1640 medium containing 10% FBS was added into the bottom chambers as chemoattractant. After incubation at 37°C for 48 h, cells inside the upper chamber were removed with cotton swabs, whereas cells on the lower membrane surface were fixed with 4% paraformaldehyde for 20 min and stained with 1% crystal violet (Sigma-Aldrich Co., St Louis, MO, USA) for 15 min. The number of invaded cells was counted in five randomly selected microscopic fields per filter under an inverted phase-contrast microscope (Olympus Corporation, Tokyo, Japan).
Western blot assay and antibodies
Cytoplasmic and nuclear proteins were lysed using the RIPA lysis buffer (Beyotime) containing protease inhibitors. The concentrations of proteins were measured using the bicinchoninic acid (BCA) assay kit (Pierce, Bonn, Germany) according to the manufacturer’s instructions. An amount of 30 μg of total protein per sample was separated by 8%–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk at room temperature and then incubated with primary antibodies overnight at 4°C. The primary antibodies used in this study included anti-zinc finger E-box-binding homeobox 2 (anti-ZEB2), anti-E-cadherin, anti-N-cadherin, anti-vimentin, and anti-GAPDH (all from Cell Signaling Technology, Beverly, MA, USA). After washing with Tris-buffered saline plus 0.1% Tween 20 (TBST), the membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology) at room temperature for 1 h. Protein bands were observed using the enhanced chemiluminescence system (Bio-Rad Laboratories Inc.). Intensity of the bands was quantified using densitometry and normalized to the GAPDH bands.
Luciferase reporter assay
The putative miRNA binding sites on SOX2OT sequences were predicted by StarBase V2.0 (http://starbase.sysu.edu.cn/) software. The SOX2OT sequences containing the predicted wild-type or mutated-type- of miR-132-binding sites were synthesized by Gene Pharma and inserted into the pmirGLO reporter vectors (Promega Corporation, Fitchburg, WI, USA) to generate the reporter vectors pmirGLO-SOX2OT-Wt (SOX2OT-Wt) and pmirGLO-SOX2T2-Mut (PVT1-Mut). For luciferase reporter assay, A549 and H1299 cells were co-transfected with luciferase plasmids with control vector containing Renilla luciferase, pRL-TK (Promega Corporation), and miR-132 mimic or miR-NC mimic. At 48 h post-transfection, luciferase activity was detected using the dual-luciferase reporter assay system (Promega Corporation).
Statistical analysis
All results were expressed as mean ± standard deviation (SD) at least from three independent experiments. Statistical analysis was performed with one-way analysis of variance (ANOVA) and Student’s t-test using the Statistical Package for the Social Sciences (SPSS) software Version 19.0 (IBM Corporation, Armonk, NY, USA). Spearman’s correlation analysis was performed to analyze correlation. A P-value of <0.05 was considered statistically significant.
Results
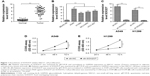
Knockdown of SOX2OT inhibits NSCLC cell proliferation
To explore SOX2OT expression levels in NSCLC tissues and cell lines, we used qRT-PCR. We found that SOX2OT expression was upregulated in NSCLC tissues compared to adjacent lung tissues (Figure 1A). Moreover, SOX2OT expression significantly increased in five NSCLC cell lines (A549, H460, H1299, NCI-H460, and HCC-827) compared to a human normal bronchus epithelial cell line, BEAS-2B (Figure 1B). To investigate the role of SOX2OT in NSCLC, we transfected sh-SOX2OT and sh-NC plasmids into A549 and H1299 cells and transfection efficiency was measured by qRT-PCR. As expected, sh-SOX2OT obviously reduced SOX2OT expression in A549 and H1299 cells (Figure 1C). Additionally, downregulation of SOX2OT by sh-SOX2OT significantly decreased the cell proliferation of A549 and H1299 cells (Figure 1D and E).
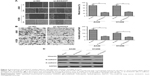
Knockdown of SOX2OT inhibits NSCLC cell migration, invasion, and EMT process
We also investigated the effect of SOX2OT on cell migration and invasion in NSCLC cells. Wound healing assay revealed that knockdown of SOXO2T significantly decreased migratory capability in both A549 and H1299 cells (Figure 2A). Transwell invasion experiments revealed that knockdown of SOX2OT apparently inhibited invasion in both A549 and H1299 cells (Figure 2B).
EMT was suggested to be crucial in the early events of tumor cell metastatic dissemination by enhancing cell migratory and invasive capacity.18,19 Thus, we hypothesized that SOX2OT promoted cell migration and invasion by inducing EMT. To test our hypothesis, we performed Western blot to examine the expression of EMT marker proteins. We found that knockdown of SOX2OT significantly increased the expression of the epithelial marker, E-cadherin, but decreased the expression of mesenchymal markers such as N-cadherin and vimentin in both A549 and H1299 cells (Figure 2C). These results suggested that SOX2OT promoted cell migration and invasion in NSCLC cells by inducing EMT.
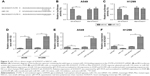
miR-132 is a direct target of SOX2OT in NSCLC cells
Since lncRNAs function as decoys of miRNAs, we predicated miRNA recognition sequences in SOX2OT using the software Starbase 2.0. We found a putative miR-132-binding site on the SOX2OT transcript (Figure 3A). To investigate if miR-132 was a functional target of SOX2OT, dual luciferase reporter assay was performed. We found that the overexpression of miR-132 significantly decreased luciferase activity of wild-type SOX2OT-3′UTR (Wt-SOX2OT) but not that of the mutant SOX2OT-3′UTR (Mut-SOX2OT) in both A549 and H1299 cells (Figure 3B and C). Next, we investigated the correlation between SOX2OT and miR-132 expression. As shown in Figure 3D, knockdown of SOX2OT increased miR-132 expression in A549 and H1299 cells. Moreover, transfection of miR-132 mimics decreased SOX2OT expression whereas transfection of miR-132 inhibitor increased SOX2OT expression in A549 and H1299 cells (Figure 3E and F). These results implied that miR-132 was a direct target of SOX2OT in NSCLC cells.
miR-132 mediates the tumor-suppressive effects of SOX2OT knockdown on NSCLC cells
To determine whether the tumor-suppressive effects of SOX2OT knockdown were mediated by miR-132 in NSCLC cells, A549 or H1299 cells were transfected with sh-SOX2OT or co-transfected with sh-SOX2OT and miR-132 inhibitor, and cell proliferation, migration, and invasion were determined. We found that co-transfection with sh-SOX2OT and miR-132 inhibitor decreased miR-132 expression in both A549 and H1299 cells compared to cells transfected with sh-SOX2OT (Figure 4A). In addition, transfection with miR-132 inhibitor partially rescued the inhibitory effect of sh-SOX2OT on cell proliferation, migration, invasion, and EMT process (Figure 4B–F). Based on the above results, we confirmed that miR-132 mediated the tumor-suppressive effects of SOX2OT knockdown in NSCLC cells.
SOX2OT controls ZEB2 expression by regulating miR-132 in NSCLC cells
A previous study has shown that miR-132 inhibited NSCLC cell migration and invasion by targeting ZEB2.20 The abovementioned results showed that miR-132 inhibitor partially mediated the tumor-suppressive effects of SOX2OT knockdown in NSCLC cells. Thus, we speculated that ZEB2 might affect SOX2OT expression in NSCLC cells. To test whether SOX2OT affected ZEB2 expression in NSCLC cells, A549 or H1299 cells were transfected with sh-SOX2OT or co-transfected with sh-SOX2OT and miR-132 inhibitor and ZEB2 expression was detected in NSCLC cells. As shown in Figure 5A and B, transfection with sh-SOX2OT significantly decreased ZEB2 expression on mRNA and protein levels, whereas co-transfection with sh-SOX2OT and miR-132 inhibitor increased ZEB2 expression levels in both A549 and H1299 cells. Moreover, we found that SOX2OT expression was negatively correlated with miR-132 expression (Figure 5C) in NSCLC tissues and positively correlated with ZEB2 expression in NSCLC tissues (Figure 5D). These results indicated that SOX2OT controlled ZEB2 expression by regulating miR-132 in NSCLC cells.
Discussion
Recently, lncRNAs have attracted attention as they appeared to be involved in tumor initiation and development in various types of cancers, including NSCLC.8,9 Many lncRNAs were confirmed to function as oncogenes or tumor suppressors in NSCLC. For instance, SNHG1 promoted NSCLC progression by modulating the miR-145-5p/MTDH axis.21 LINC00460 acted as an oncogene in NSCLC to promote cell migration and invasion by inducing EMT.22 lncRNA XIST increased cell viability and invasion by regulating the miR-137/PXN axis in NSCLC.23 lncRNA MEG3 suppressed proliferation and induced apoptosis by affecting p53 expression in NSCLC cells.24 In the present study, we examined SOX2OT expression levels in NSCLC samples from 48 patients with NSCLC and five NSCLC cell lines and found that SOX2OT expression was significantly upregulated in NSCLC tissues and cell lines, suggesting that SOX2OT functioned as oncogene in NSCLC. This oncogene function of SOX2OT was further examined by using NSCLC cell lines, knocking down SOX2OT. The results demonstrated that knockdown of SOX2OT in A549 and H1299 cells significantly decreased cell proliferation, migration, and invasion and inhibited the EMT process. Mechanistic study revealed that SOX2OT exerted oncogene role in NSCLC by regulating miR-132. Thus, these results highlight a novel SOX2OT/miR-132 axis in regulating NSCLC cell migration, invasion, and EMT process.
SOX2OT, localized on human chromosome 3q26.33, was highly expressed in multiple types of cancers.10–16 Recent studies have reported that SOX2OT levels were higher in NSCLC tissues than those in healthy controls, and SOX2OT served as a promising biomarker for diagnosing and monitoring NSCLC.17,25 In agreement with the results of previous studies, we found that SOX2OT was upregulated in NSCLC tissues and cell lines compared to normal lung tissues and a human bronchial epithelial cell line. A report by Hou et al17 has shown that SOX2OT promoted NSCLC cell proliferation by regulating cell arrest at the G2/M phase.17 However, its role in migration and invasion of NSCLC has not been well studied. The present study showed that knockdown of SOX2OT inhibited NSCLC cell migration and invasion by inhibiting EMT process. In accordance with previous results, our results suggested that SOX2OT functioned as oncogene in NSCLC cells.
It was well known that lncRNAs could serve as competing endogenous RNAs (ceRNAs) or as molecular sponges in modulating the concentration and biological functions of miRNAs.26 SOX2OT has been identified as ceRNA in several cancers by sponging miRNAs, including miR-375,27 miR-211,15 miR-194-5p, and miR-122.11 In the present study, bioinformatics analysis demonstrated a putative binding site in SOX2OT for miR-132. Dual luciferase reporter assays further confirmed the direct binding between miR-132 and SOX2OT. miR-132 functions as a tumor suppressor, which inhibits NSCLC cell proliferation, migration, invasion, and EMT process.28–30 In this study, we demonstrated that knockdown of SOX2OT increased miR-132 expression in both A549 and H1299 cells. Moreover, transfection of miR-132 mimics decreased SOX2OT expression, whereas transfection of miR-132 inhibitor increased SOX2OT expression in A549 and H1299 cells. Interestingly, an inverse relationship between miR-132 and SOX2OT was observed in NSCLC tissues. We also demonstrated that transfection with miR-132 inhibitor rescued the inhibitory effect of sh-SOX2OT on cell proliferation, migration, invasion, and EMT process. These results implied that miR-132 mediated the tumor-suppressive effects of SOX2OT knockdown in NSCLC cells.
Aforementioned results showed that SOX2OT elicited its oncogene function by acting as a sponge for miR-132 in NSCLC cells, which might affect the ability of miR-132 to bind its targets. A previous study has demonstrated that miR-132 inhibited cell migration and invasion by targeting ZEB2.20 ZEB2, an E-box-binding transcription factor, was involved in the tumorigenesis of various malignancies.31 Moreover, ZEB2 was reported to promote tumor cell invasion and metastasis as it could directly bind the E-cadherin promoter and strongly inhibit E-cadherin expression during EMT.32 Here, we found that knockdown of SOX2OT significantly decreased ZEB2 expression on mRNA and protein levels, whereas co-transfection with sh-SOX2OT and miR-132 inhibitor increased ZEB2 expression levels in both A549 and H1299 cells. Moreover, we found that SOX2OT expression was negatively correlated with miR-132 expression in NSCLC tissues and positively correlated with ZEB2 expression in NSCLC tissues. There results suggested that SOX2OT exerted its oncogene function, at least in part, by regulating the miR-132/ZEB2 axis.
Conclusion
The present study indicated that SOX2OT expression was upregulated in NSCLC tissues and cell lines. Function assays demonstrated that SOX2OT promoted cell migration and invasion by inducing EMT in NSCLC cells. Mechanistic study implied that SOX2OT exerted an oncogene role in NSCLC cells by regulating the miR-132/ZEB2 axis. Hence, our study elucidates the function and molecular mechanism of SOX2OT in NSCLC cells and highlights its therapeutic values for NSCLC patients.
Acknowledgment
This study was supported by the grants from National Natural Science Foundation of China (81401883).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Carbone DP, Felip E. Adjuvant therapy in non-small cell lung cancer: future treatment prospects and paradigms. Clin Lung Cancer. 2011;12(5):261–271. | ||
Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. | ||
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. | ||
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. | ||
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | ||
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. | ||
Wei MM, Zhou GB. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14(5):280–288. | ||
Xie W, Yuan S, Sun Z, Li Y. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics. 2016;8(9):1275–1287. | ||
Han L, Zhang W, Zhang B, Zhan L. Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Exp Ther Med. 2018;15(2):2182–2188. | ||
Su R, Cao S, Ma J, et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer. 2017;16(1):171. | ||
Tang X, Gao Y, Yu L, et al. Correlations between lncRNA-SOX2OT polymorphism and susceptibility to breast cancer in a Chinese population. Biomark Med. 2017;11(3):277–284. | ||
Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8(11):5035–5043. | ||
Liu S, Xu B, Yan D. Enhanced expression of long non-coding RNA Sox2ot promoted cell proliferation and motility in colorectal cancer. Minerva Med. 2016;107(5):279–286. | ||
Shafiee M, Aleyasin SA, Vasei M, Semnani SS, Mowla SJ. Down-regulatory effects of miR-211 on long non-coding RNA SOX2OT and SOX2 genes in esophageal squamous cell carcinoma. Cell J. 2016;17(4):593–600. | ||
Shi XM, Teng F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol. 2015;8(4):4008–4014. | ||
Hou Z, Zhao W, Zhou J, et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388. | ||
Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. | ||
Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22(6):699–701. | ||
You J, Li Y, Fang N, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One. 2014;9(3):e91827. | ||
Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32(7):3957–3967. | ||
Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. | ||
Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. | ||
Lu KH, Li W, Liu XH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. | ||
Xie Y, Zhang Y, Du L, et al. Circulating long non-coding RNAs act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol. 2018;12(5):648–658. | ||
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. | ||
Shafiee M, Aleyasin SA, Mowla SJ, Vasei M, Yazdanparast SA. The effect of MicroRNA-375 overexpression, an inhibitor of helicobacter pylori-induced carcinogenesis, on lncRNA SOX2OT. Jundishapur J Microbiol. 2016;9(9):e23464. | ||
Guo H, Zhang X, Chen Q, Bao Y, Dong C, Wang X. miR-132 suppresses the migration and invasion of lung cancer cells by blocking USP9X-induced epithelial-mesenchymal transition. Am J Transl Res. 2018;10(1):224–234. | ||
Zhang JX, Zhai JF, Yang XT, Wang J. MicroRNA-132 inhibits migration, invasion and epithelial-mesenchymal transition by regulating TGFbeta1/Smad2 in human non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20(18):3793–3801. | ||
Li Y, Zu L, Wang Y, Wang M, Chen P, Zhou Q. miR-132 inhibits lung cancer cell migration and invasion by targeting SOX4. J Thorac Dis. 2015;7(9):1563–1569. | ||
Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132(4):745–754. | ||
Wong TS, Gao W, Chan JY. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. Biomed Res Int. 2014;2014:921564. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.