Back to Journals » OncoTargets and Therapy » Volume 13
Long Noncoding RNA RGMB-AS1 Acts as a microRNA-574 Sponge Thereby Enhancing the Aggressiveness of Gastric Cancer via HDAC4 Upregulation
Authors Wang X, Chen X, Tian Y, Jiang D, Song Y
Received 10 October 2019
Accepted for publication 11 January 2020
Published 24 February 2020 Volume 2020:13 Pages 1691—1704
DOI https://doi.org/10.2147/OTT.S234144
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper is retracted.
Xiaodong Wang, Xin Chen, Yueli Tian, Dongqiang Jiang, Ying Song
Department of Gastroenterology and Digestive Endoscopy Center, The Second Hospital of Jilin University, Changchun, Jilin 130041, People’s Republic of China
Correspondence: Ying Song
Department of Gastroenterology and Digestive Endoscopy Center, The Second Hospital of Jilin University, 218 Ziqiang Road, Changchun, Jilin 130041, People’s Republic of China
Email [email protected]
Purpose: The long noncoding RNA RGMB-AS1 plays an important part in the genesis and progression of multiple human cancers. Nonetheless, little is known regarding its expression, roles, and mechanisms of action in gastric cancer (GC). This study was aimed at investigating the relationship between RGMB-AS1 and GC and illustrating the mechanisms of action of RGMB-AS1 therein.
Methods: RGMB-AS1 expression in GC was measured via reverse-transcription quantitative PCR. A series of experiments including Cell Counting Kit-8 assay, flow-cytometric analysis of apoptosis, Transwell migration and invasion assays, and in vivo tumorigenesis experiment were conducted to test the effects of RGMB-AS1 on the malignant phenotype of GC cells. The molecular events behind the oncogenic actions of RGMB-AS1 in GC were elucidated through subcellular fractionation, RNA immunoprecipitation assay, bioinformatics analysis and luciferase reporter assay.
Results: RGMB-AS1 upregulation was confirmed in GC tissues and cell lines. Higher RGMB-AS1 expression was associated with adverse clinical parameters and negatively correlated with patient overall survival. RGMB-AS1 knockdown inhibited GC cell proliferation, facilitated apoptosis, and reduced migration and invasion in vitro. Further experiments revealed that RGMB-AS1 knockdown decreased the tumor growth of GC cells in vivo. Mechanistically, RGMB-AS1 functioned as a competing endogenous RNA upregulating histone deacetylase 4 (HDAC4) by sponging microRNA-574 (miR-574). Rescue experiments indicated that miR-574 inhibition and HDAC4 reintroduction reversed the effects of the RGMB-AS1 knockdown on GC cells.
Conclusion: The RGMB-AS1–miR-574–HDAC4 regulatory network contributes to the malignancy of GC, thereby offering a novel target for the diagnosis, prognosis, and/or treatment of GC.
Keywords: tumor therapy, gastric cancer, miR-574, RGMB-AS1, histone deacetylase 4
Introduction
Gastric cancer (GC), which originates in the gastric mucosa, is the fourth most prevalent human cancer and its mortality rate ranks second among cancer-associated mortality rates globally.1,2 Approximately 1.033 million new GC cases and 783 000 GC-related deaths worldwide are estimated for 2018, according to 2018 GLOBOCAN statistics.2 Despite the tremendous achievements in GC diagnosis and therapy, its treatment efficacy at an advanced stage remains poor, with a 5-year overall survival rate of only approximately 20–30%.3 These poor clinical outcomes are attributable to multiple factors including a lack of early diagnosis in addition to recurrence and local/distant metastasis even following surgical resection.4 Although the incidence of GC is closely related to Helicobacter pylori infection, diet, smoking, and obesity,5 its pathogenesis is complicated and the mechanisms underlying GC initiation and progression remain largely unknown. Hence, it is critical to elucidate the mechanisms responsible for the malignancy of GC and to identify novel effective targets applicable to the diagnosis and therapy of GC, with the aim of improving patient prognosis.
Long noncoding RNAs (lncRNAs) are a series of recently discovered RNA molecules greater than 200 nucleotides in length.6 These transcripts do not encode proteins but are implicated in the modulation of gene expression at transcriptional and post-transcriptional levels.7 Previously, lncRNAs have been regarded as “noise” in genome transcription; nevertheless, increasing numbers of studies have revealed their contribution to nearly all parameters of biological and pathological processes.8–10 Substantial research has demonstrated an alteration of lncRNA expression in GC, with some of the identified lncRNAs performing a vital function in regulating gastric carcinogenesis and GC progression.11–13 Notably, lncRNAs may serve as either tumor suppressors or oncogenic molecules and participate in the regulation of malignant characteristics of GC during GC progression.14–16
MicroRNAs (miRNAs) comprise another group of noncoding RNA molecules, of 17–25 nucleotides in length.17 They can directly interact with the 3′ untranslated regions (3′-UTRs) of their target mRNAs in a base-pairing manner, thereby causing mRNA degradation and/or translational suppression. A variety of mRNAs have also been found to be abnormally expressed in human cancers, such as GC. The dysregulation of miRNAs in GC has been functionally characterized and found to exert important actions on multiple pathophysiological parameters. Increasing evidence shows that lncRNAs act as competing endogenous RNAs (ceRNAs) toward specific miRNAs and thereby positively modulate the expression of these mRNAs. Consequently, the resultant lncRNA–miRNA–mRNA regulatory network may become a diagnostic biomarker and therapeutic target in GC.
LncRNA RGMB-AS1 performs an important function in the genesis and progression of multiple human cancers.18–21 Nevertheless, the expression and functions of RGMB-AS1 in GC remained unknown. Therefore, this study was designed to determine the expression level and detailed involvement of RGMB-AS1 in GC. Moreover, we thoroughly examined the mechanism by which RGMB-AS1 promotes GC progression.
Materials and Methods
Collection of Tissue Samples
The study protocol was approved by the Ethics Committee of The Second Hospital of Jilin University. All the participants signed an informed consent form. Tumor tissue samples and adjacent normal tissues were obtained in The Second Hospital of Jilin University from 67 patients with GC who had not undergone preoperative radiotherapy, chemotherapy, or other anticancer treatments. None of these patients had a diagnosis of other types of cancer. Following surgical resection, all fresh tissue samples were quickly immersed in liquid nitrogen and then stored at −80 °C.
Cell Lines
A normal human gastric epithelial cell line (GES-1) and four human GC cell lines (MKN-45, MGC-803, BGC-823, and AGS) were purchased from the American Type Culture Collection (Manassas, VA, USA). A culture medium consisting of Dulbecco’s modified Eagle’s medium (DMEM; cat. No.12491-015), 10% fetal bovine serum (FBS; cat. No.10091148), 100 U/mL penicillin, and 100 μg/mL streptomycin (cat. No.15070063; all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was utilized for cell cultivation. All cells were kept at 37 °C in an incubator containing an atmosphere of humidified 95% air and 5% CO2.
Oligonucleotides, Plasmids, and Cell Transfection
The small interfering RNAs (siRNAs) targeting RGMB-AS1 (si-RGMB-AS1) and negative control siRNA (si-NC) were synthesized by RiboBio Co., Ltd. (Guangzhou, China). The si-RGMB-AS1 sequence was 5ʹ- GGCTCAATTACCGACACAGTTCAAG-3ʹ and the si-NC sequence was 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ. The agomirs for miR-574 (agomir-574 and agomir-NC), and miRNA antagomirs (antagomir-574 and antagomir-NC) were all purchased from GenePharma Co., Ltd. (Shanghai, China). The agomir-574 sequence was 5ʹ-CACGCUCAUGCACACACCCACA-3ʹ, the agomir-NC sequence was 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ, antagomir-574 sequence was 5ʹ-UGUGGGUGUGUGCAUGAGCGUG −3ʹ, and the antagomir-NC sequence was 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ. HDAC4 overexpression plasmid pcDNA3.1-HDAC4 (hereafter: pc-HDAC4), was chemically synthesized by Generay (Shanghai, China), and the empty pcDNA3.1 vector served as the control.
Cells growing in the logarithmic (log) phase were seeded in 6-well plates and cultured at 37 °C overnight. All above-mentioned siRNAs, agomirs, antagomirs, and plasmids were introduced into cells using Lipofectamine™ 2000 (cat. No.11668019; Invitrogen; Thermo Fisher Scientific) in accordance with the relevant protocols.
RNA Extraction and Reverse-Transcription Quantitative PCR (RT-qPCR)
The TRIzol® reagent (cat. No.15596026; Invitrogen; Thermo Fisher Scientific) was employed for isolation of total RNA from tissues or cells. After quality verification on a NanoDrop™ 2000 spectrophotometer (Invitrogen; Thermo Fisher Scientific), total RNA was reverse-transcribed into complementary DNA (cDNA) by means of the TaqMan MicroRNA Reverse Transcription Kit (cat. No. 4366596; Applied Biosystems, Foster City, CA, USA), and the obtained cDNA was then subjected to qPCR for the measurement of miR-574 expression using the TaqMan MicroRNA Assay Kit (cat. No. 4427975; Applied Biosystems). MiR-574 expression was normalized to that of U6 small nuclear RNA. To determine RGMB-AS1 and HDAC4 expression, reverse transcription was conducted using the PrimeScript RT Reagent Kit (cat. No. RR037A; TaKaRa Biotechnology Co., Ltd., Dalian, China), after which qPCR was performed by means of the SYBR Premix Ex Taq™ kit (cat. No. RR420A; TaKaRa Biotechnology). GAPDH served as the internal reference for RGMB-AS1 and HDAC4.
The primers were designed as follows: RGMB-AS1, 5′-AGTGGGCAAACTTCAACGTTC-3′ (forward) and 5′-GAGCTGCCATTGAATTAATCCG-3′ (reverse); HDAC4, 5′-AGAATGGCTTTGCTGTGGTC-3′ (forward) and 5′-ATCTTGCTCACGCTCAACCT-3′ (reverse); and GAPDH, 5′-GAGTCCACTGGCGTCTTC-3′ (forward) and 5′- GATGATCTTGAGGCTGTTGTC-3′ (reverse). Relative gene expression was analyzed using the 2–ΔΔCq method.
Cell Counting Kit-8 (CCK-8) Assay
At 24 h post-transfection, cells were seeded in 96-well plates at a density of 2 × 103 cells per well. Cell proliferation was evaluated by the addition of 10 μL of the CCK-8 solution (cat. No. CK04; Dojindo Laboratories, Kumamoto, Japan) into each well. After an additional 2 h incubation, absorbance was measured at 450 nm wavelength using a Microplate Spectrophotometer (BioTek, Winooski, VT, USA). The CCK-8 assay was carried out at four time points (0, 24, 48, and 72 h after seeding), and the growth curves were plotted with time points on the X-axis and absorbance values on the Y-axis.
Flow-Cytometric Analysis of Apoptosis
The apoptotic status of GC cells was tested using the Annexin V Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (cat. No. 640914; Biolegend, San Diego, CA, USA). In particular, transfected cells were harvested with EDTA-free 0.25% trypsin, rinsed twice with ice-cold phosphate-buffered saline, centrifuged at 4 °C, then the supernatant was decanted. The transfected cells were resuspended in 100 µL of 1× binding buffer and 5 µL Annexin V-FITC and 5 µL propidium iodide solution were added for double staining of apoptotic cells. After 15 min incubation in the dark, the percentage of apoptotic cells was determined on a FACScan flow cytometer.
Transwell Migration and Invasion Assays
To evaluate the effects of the miRNA on cellular invasive capacity, 24-well insert Transwell chambers (cat. No. 353093) coated with Matrigel (cat. No. 356231; both from BD Biosciences, San Jose, CA, USA) were applied for the Transwell invasion assay. Transfected cells were harvested through digestion with 0.25% trypsin and rinsed twice with phosphate-buffered saline. After centrifugation, the supernatant was discarded and the transfected cells were resuspended in serum-free DMEM. A total of 100 µL of the suspension containing 5 × 104 cells was transferred to the upper compartment of each chamber, whereas 600 µL of DMEM supplemented with 20% FBS was placed in the bottom compartments. After cultivation for 24 h at 37 °C, the cells remaining in the upper chamber were carefully removed with a cotton swab, whereas the invasive cells were fixed with 4% polyformaldehyde and stained with 0.5% crystal violet. The stained cells were imaged and counted under an inverted microscope (IX83; Olympus, Tokyo, Japan). Similar experimental procedures were conducted to determine the cellular migratory ability, except that the chambers were not precoated with Matrigel.
In vivo Tumorigenesis Experiment
Animal maintenance and experimental steps were approved by the Animal Care and Use Committee of The Second Hospital of Jilin University. All experimental steps were performed in compliance with the Animal Protection Law of the People’s Republic of China-2009 for experimental animals. Female nude mice at the age of 4–6 weeks were purchased from Shanghai Laboratory Animal Center (Shanghai, China). Cells transfected with either si-RGMB-AS1 or si-NC were harvested at 24 h of cultivation and injected subcutaneously into a flank of the mice. Measurements of the width and length of the resultant tumor xenografts were carried out every five days, and the volume was calculated via the following formula: 0.5 × length × (width)2. The tumor xenografts were allowed to grow in vivo for 30 days. On day 30, all the mice were euthanized by cervical dislocation and the tumor xenografts were excised, weighed, and subjected to RT-qPCR and Western blotting analyses.
Subcellular Fractionation
The PARIS Kit (cat. No. AM1921; Invitrogen; Thermo Fisher Scientific) was employed to collect and separate cytoplasmic and nuclear fractions of GC cells. RNA was isolated from the cytoplasmic and nuclear fractions using the TRIzol® reagent and subjected to RT-qPCR analysis to evaluate RGMB-AS1 distribution within GC cells.
RNA Immunoprecipitation (RIP) Assay
The Magna RNA-binding Protein Immunoprecipitation Kit (cat. No. 17-701; Millipore, Billerica, MA, USA) was utilized to perform the RIP assay. Cells were rinsed thrice with phosphate-buffered saline, trypsinized, and treated with RIP lysis buffer. The cell extracts were collected and incubated with magnetic beads conjugated with human anti-Argonaute2 (AGO2) or anti-IgG antibodies (Millipore). The magnetic beads were washed and then incubated with proteinase K in order to digest proteins. Finally, RT-qPCR was carried out for quantification of RGMB-AS1 and miR-574 in the coprecipitated RNA samples.
Bioinformatics Analysis and Luciferase Reporter Assay
The interaction between RGMB-AS1 and miRNA was predicted using starBase 3.0 software (http://starbase.sysu.edu.cn/). TargetScan (http://www.targetscan.org) and starBase 3.0 were applied to predict the putative targets of miR-574. Wild-type (wt) RGMB-AS1 that contains a predicted miR-574–binding sequence and mutant (mut) RGMB-AS1 were chemically synthesized by GenePharma Co., Ltd. and integrated into the pMIR-REPORT plasmid (cat. No. AM5795; Invitrogen; Thermo Fisher Scientific) to respectively generate reporter plasmids pMIR-wt-RGMB-AS1 and pMIR-mut-RGMB-AS1. The reporter plasmids intended for determining the interaction between miR-574 and HDAC4 mRNA, namely, pMIR-wt-HDAC4 and pMIR-mut-HDAC4, were constructed via similar experimental steps. For the reporter assay, cotransfection of either the wt or mut reporter plasmid and either agomir-574 or agomir-NC into GC cells was performed using Lipofectamine™ 2000. After 48 h cultivation, the Dual Luciferase Reporter Assay Kit (cat. No. E1910; Promega, Madison, WI, USA) was used to quantify the luciferase activity. Renilla luciferase activity served as the control for firefly luciferase activity.
Protein Isolation and Western Blotting
Transfected MGC-803 and AGS cells were lysed with RIPA buffer (cat. No. P0013B; Beyotime Institute of Biotechnology, Shanghai, China). The Bicinchoninic Acid Protein Assay Kit (cat. No. P0009; Beyotime Institute of Biotechnology) was utilized for the quantification of total protein. Identical amounts of protein were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Prior to incubation with primary antibodies overnight at 4 °C, the membranes were blocked with 5% nonfat dry milk at room temperature for 2 h. Next, after incubation with horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (cat. No. ab205719; dilution 1:1000; Abcam, Cambridge, UK), the SuperSignal West Femto Maximum Sensitivity Substrate (cat. No.34096; Thermo Fisher Scientific) was applied to visualize the protein signals on the membranes. The anti-HDAC4 antibody (cat. No. ab234084; dilution 1:1000; Abcam) was used herein and GAPDH (cat. No. ab9484; dilution 1:1000; Abcam) served as a loading control.
Statistical Analysis
Data are presented as the mean ± standard deviation. The Chi-squared test was performed to determine the relationship between RGMB-AS1 and clinical parameters among the patients with GC. Student’s t test was conducted to analyze the differences between two groups. Differences among multiple groups were evaluated by one-way analysis of variance followed by Tukey’s test. Survival curves were plotted using the Kaplan–Meier method and analyzed by the log rank test. The expression correlation between RGMB-AS1 and miR-574 among the GC tissue samples was assessed by Spearman correlation analysis. MiR-574 and HDAC4 expression correlation was also evaluated via Spearman correlation analysis. P < 0.05 was assumed to indicate a statistically significant difference.
Results
High Expression of RGMB-AS1 Correlates with Adverse Clinical Parameters and Worse Clinical Outcomes of GC
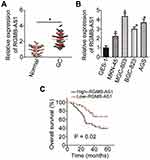
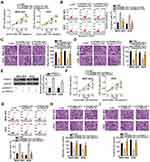
First, we measured the expression of RGMB-AS1 in 67 pairs of GC tissue samples and adjacent normal tissues. The results of RT-qPCR analysis indicated that RGMB-AS1 expression was higher in GC tumors than that in adjacent normal tissues (Figure 1A, P < 0.05). We also measured RGMB-AS1 expression in four GC cell lines (MKN-45, MGC-803, BGC-823, and AGS) and in normal human gastric epithelial cells (GES-1) via RT-qPCR. All four tested GC cell lines manifested higher expression of RGMB-AS1 than that in GES-1 cells (Figure 1B, P < 0.05).
According to the median value of RGMB-AS1 among the GC tissue samples, all 67 patients with GC were classified into high- or low-RGMB-AS1 expression groups, and then the association between RGMB-AS1 expression and clinical parameters among the patients with GC was assessed. An elevated RGMB-AS1 level closely correlated with tumor size (P = 0.026), TNM stage (P = 0.043), and lymph node metastasis (P = 0.029; Table 1). Furthermore, Kaplan–Meier analysis along with the log rank test was conducted to evaluate whether the expression of RGMB-AS1 correlated with overall survival of the patients under study. Patients with GC in the high-RGMB-AS1 expression group exhibited shorter overall survival than those in the low-RGMB-AS1 expression group (Figure 1C, P = 0.02). Overall, RGMB-AS1 was found to be aberrantly overexpressed in GC and this aberration may be involved in GC progression.
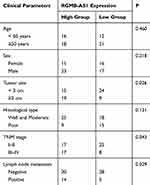 |
Table 1 Associations Between RGMB-AS1 Expression and Clinical Parameters of Patients with GC (n=67) |
Knockdown of RGMB-AS1 Restricts the Proliferation, Migration, and Invasion and Facilitates the Apoptosis of GC Cells in vitro
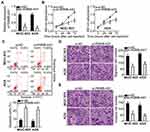
To study the effects of RGMB-AS1 on the GC tumor phenotype, si-RGMB-AS1 was synthesized and transfected into MGC-803 and AGS cells. RT-qPCR analysis verified the successful knockdown of RGMB-AS1 in both cell types (Figure 2A, P < 0.05). MGC-803 and AGS cells were also transfected with or without si-NC. After transfection, expression of RGMB-AS1 was detected by means of RT-qPCR. Transfection with si-NC did not affect the expression of RGMB-AS1 (Supplementary Figure 1A). Next, the CCK-8 assay was performed to evaluate cellular proliferation. The results suggested that transfection with si-RGMB-AS1 led to evident suppression of MGC-803 and AGS cell proliferation (Figure 2B, P < 0.05). In addition, our results revealed that the knockdown of RGMB-AS1 markedly promoted the apoptosis of MGC-803 and AGS cells (Figure 2C, P < 0.05), as evidenced by the flow-cytometric analysis. Furthermore, Transwell cell migration and invasion assays were conducted to test whether RGMB-AS1 is involved in GC cell metastasis in vitro. A reduction in RGMB-AS1 expression obviously impaired the migratory (Figure 2D, P < 0.05) and invasive abilities (Figure 2E, P < 0.05) of MGC-803 and AGS cells. Collectively, these results implied that RGMB-AS1 performs oncogenic activities during the growth and metastasis of GC cells in vitro.
RGMB-AS1 Acts as a ceRNA on miR-574 in GC Cells
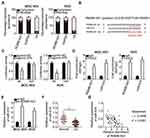
Some studies have shown that lncRNAs work as ceRNAs for specific miRNAs to modulate their expression and functions. To elucidate the mechanisms behind the oncogenic actions of RGMB-AS1 in GC, subcellular localization of RGMB-AS1 was first characterized in MGC-803 and AGS cells. The results indicated that the majority of RGMB-AS1 was located in the cytoplasm of MGC-803 and AGS cells (Figure 3A), indicating that this lncRNA may regulate its target at the post-transcriptional levels. Bioinformatics analysis revealed that miR-574, a GC-related tumor suppressor,22,23 might constitute a potential target of RGMB-AS1 (Figure 3B). To validate this prediction, a luciferase reporter assay was conducted to evaluate the binding of RGMB-AS1 to miR-574 in GC cells. Either plasmid pMIR-wt-RGMB-AS1 or pMIR-mut-RGMB-AS1 was introduced into MGC-803 and AGS cells with either agomir-574 or agomir-NC. Cotransfection with pMIR-wt-RGMB-AS1 and agomir-574 resulted in an obvious decrease in luciferase activity in MGC-803 and AGS cells (P < 0.05), whereas the luciferase activity remained unaltered when these cells were cotransfected with pMIR-mut-RGMB-AS1 (Figure 3C). In addition, the direct interaction between RGMB-AS1 and miR-574 was examined using the RIP assay. The results revealed that miR-574 and RGMB-AS1 from lysates of MGC-803 and AGS cells were significantly enriched on AGO2-bound beads compared with the IgG control (Figure 3D, P < 0.05).
Next, we assessed the expression of miR-574 in RGMB-AS1-deficient MGC-803 and AGS cells. RT-qPCR analysis confirmed that the RGMB-AS1 knockdown obviously enhanced the expression of miR-574 in MGC-803 and AGS cells (Figure 3E, P < 0.05). In addition, we tested the expression correlation between miR-574 and RGMB-AS1 in GC tissue samples. Consistent with the results of other studies,22,23 miR-574 was markedly downregulated in the GC tissue samples relative to the levels in adjacent normal tissues (Figure 3F, P < 0.05). Furthermore, the expression of miR-574 manifested an inverse correlation with RGMB-AS1 expression in the 67 GC tissue samples (Figure 3G; r = –0.5446, P < 0.0001). In summary, RGMB-AS1 was found to function as a ceRNA for miR-574 in GC cells.
MiR-574 Inhibits the Malignant Properties of GC Cells in vitro and Directly Targets HDAC4 mRNA in GC Cells
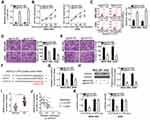
To explore the biological roles of miR-574 in GC, either agomir-574 or agomir-NC was transfected into GC cells and the influence of miR-574 upregulation on the malignant properties of GC cells was investigated. First, the upregulation of miR-574 in agomir-574–transfected MGC-803 and AGS cells was verified by RT-qPCR (Figure 4A, P < 0.05). Transfection with agomir-574 significantly decreased MGC-803 and AGS cell proliferation (Figure 4B, P < 0.05), increased apoptosis (Figure 4C, P < 0.05), and impaired migration (Figure 4D, P < 0.05) and invasion (Figure 4E, P < 0.05) in vitro. Collectively, these results confirmed that miR-574 exerts a tumor-suppressive action on the malignancy of GC cells in vitro.
We next attempted to uncover the mechanism of action of this miRNA. Bioinformatics analysis suggested that HDAC4 contains highly conserved binding sequences for miR-574 (Figure 4F). To test whether miR-574 is implicated in the modulation of HDAC4 expression, RT-qPCR and Western blotting were performed to respectively measure HDAC4 mRNA and protein expression in MGC-803 and AGS cells following either agomir-574 or agomir-NC transfection. The results showed that upregulation of miR-574 markedly decreased HDAC4 mRNA (Figure 4G, P < 0.05) and protein (Figure 4H, P < 0.05) levels in both cell types. In addition, HDAC4 mRNA expression in GC tissue samples and adjacent normal tissues (n = 67) was assessed by RT-qPCR. The expression of HDAC4 mRNA was considerably higher in the GC tissue samples (Figure 4I, P < 0.05), manifesting a negative correlation with miR-574 expression (Figure 4J; r = –0.5747, P < 0.0001). Finally, the luciferase reporter assay was carried out to ascertain whether miR-574 can bind directly to HDAC4 mRNA. miR-574 overexpression decreased the luciferase activity generated by plasmid pMIR-wt-HDAC4 (P < 0.05) in MGC-803 and AGS cells. In contrast, the luciferase activity generated by plasmid pMIR-mut-HDAC4 remained unaltered following agomir-574 cotransfection (Figure 4K). These data provided evidence that HDAC4 is a direct target gene of miR-574 in GC cells.
RGMB-AS1 Sponges miR-574 to Increase HDAC4 Expression in GC Cells
We next aimed to test whether HDAC4 expression can be modulated by RGMB-AS1 via the sponging of miR-574. Hence, either si-RGMB-AS1 or si-NC was transfected into MGC-803 and AGS cells, and HDAC4 expression was quantified at the mRNA and protein levels. The results indicated that the mRNA (Figure 5A, P < 0.05) and protein levels (Figure 5B, P < 0.05) of HDAC4 were decreased by RGMB-AS1 knockdown in MGC-803 and AGS cells. In addition, MGC-803 and AGS cells were transfected with or without si-NC. Expression levels of miR-574 and RGMB-AS1 protein were respectively detected by RT-qPCR and Western blotting. The results displayed that transfection with si-NC did not alter the expression of miR-574 and RGMB-AS1 protein compared with that in cells transfected without si-NC (Supplementary Figure 1B and C)
Furthermore, the expression of HDAC4 was analyzed in MGC-803 and AGS cells following cotransfection with si-RGMB-AS1 and either antagomir-NC or antagomir-574. First, RT-qPCR analysis confirmed that the transfection with antagomir-574 successfully reduced miR-574 expression in both cell types (Figure 5C, P < 0.05). Notably, the decrease in HDAC4 mRNA (Figure 5D P < 0.05) and protein amounts (Figure 5E, P < 0.05) under the influence of RGMB-AS1 knockdown was reversed by cotransfection of MGC-803 and AGS cells with antagomir-574. These results implied that RGMB-AS1 serves as a ceRNA that increases HDAC4 expression in GC cells by competing for miR-574.
The RGMB-AS1–miR-574–HDAC4 Pathway Promotes GC Progression in vitro
Rescue assays were carried out to identify the functions of the RGMB-AS1–miR-574–HDAC4 pathway in the malignancy of GC cells. Toward this end, either antagomir-574 or antagomir-NC along with si-RGMB-AS1 were transfected into MGC-803 and AGS cells. The CCK-8 assay and flow-cytometric analysis of apoptosis confirmed that the suppression of cell proliferation (Figure 6A, P < 0.05) and promotion of apoptosis (Figure 6B, P < 0.05) by the RGMB-AS1 knockdown were reversed by antagomir-574 cotransfection into MGC-803 and AGS cells. Transwell migration and invasion assays indicated that cell migratory (Figure 6C, P < 0.05) and invasive abilities (Figure 6D, P < 0.05) were impaired by RGMB-AS1 knockdown albeit recovered in MGC-803 and AGS cells by means of antagomir-574 cotransfection.
In addition, rescue experiments were performed on MGC-803 and AGS cells following cotransfection of si-RGMB-AS1 and either pcDNA3.1 or pc-HDAC4. The transfection efficiency is shown in Figure 6E (P < 0.05). As anticipated, the effects of RGMB-AS1 knockdown on GC cell proliferation (Figure 6F, P < 0.05), apoptosis (Figure 6G, P < 0.05), migration (Figure 6H, P < 0.05), and invasion (Figure 6I, P < 0.05) were attenuated by HDAC4 reintroduction. Overall, the above results confirmed that knockdown of RGMB-AS1 decreased GC progression via the miR-574–HDAC4 axis.
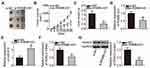
RGMB-AS1 Knockdown Inhibits Tumor Growth of GC Cells in vivo
MGC-803 cells transfected with either si-RGMB-AS1 or si-NC were inoculated into the flank of nude mice to test the influence of RGMB-AS1 on the tumor growth of GC cells in vivo. It was observed that the volume (Figure 7A and B, P < 0.05) and weight (Figure 7C, P < 0.05) of the tumor xenografts derived from si-RGMB-AS1-transfected MGC-803 cells were significantly lower than those of xenografts formed by si-NC–transfected cells. In addition, a decrease in RGMB-AS1 (Figure 7D, P < 0.05) and an increase in miR-574 (Figure 7E, P < 0.05) amounts were noted in the tumor xenografts from the si-RGMB-AS1 group. Furthermore, HDAC4 mRNA (Figure 7F, P < 0.05) and protein levels (Figure 7G, P < 0.05) in the tumor xenografts derived from MGC-803 cells transfected with si-RGMB-AS1 were lower than those in the si-NC group. Taken together, these findings indicated that a reduction in RGMB-AS1 expression lowered HDAC4 expression through decreased sponging of miR-574 in GC cells, resulting in the inhibition of tumor growth in vivo.
Discussion
Recently, increasing evidence indicates that a large number of lncRNAs are dysregulated in GC.24–26 The dysregulation of lncRNAs is implicated in the regulation of various malignant cytological behaviors during gastric carcinogenesis and GC progression.27–29 Therefore, comprehensive research into the lncRNA-based regulatory network in GC may facilitate the discovery of effective targets for the diagnosis and treatment of this cancer. Although a variety of lncRNAs in GC have been well-studied, numerous other lncRNAs remain to be further explored in detail. In the present study, a novel GC-specific lncRNA, RGMB-AS1, was identified. We measured its expression profile, determined detailed functions, and illustrated the mechanism of action of RGMB-AS1 in GC.
RGMB-AS1 is overexpressed in laryngeal squamous cell carcinoma and shows a significant correlation with tumor stage and lymph node metastasis.18 Patients with laryngeal squamous cell carcinoma showing high RGMB-AS1 expression exhibit worse overall-survival and disease-free survival rates than those of patients with low RGMB-AS1 expression.18 RGMB-AS1 is upregulated in lung cancer19 and papillary thyroid carcinoma.20 RGMB-AS1 upregulation in lung cancer is related to differentiation, TNM stage, and lymph node metastasis.19 In contrast, RGMB-AS1 is downregulated in hepatocellular carcinoma and its downregulation shows an obvious association with clinical stage, tumor size, and metastasis.21 In addition, RGMB-AS1 has been validated as an independent predictor of favorable clinical outcomes of patients with hepatocellular carcinoma.21 Nonetheless, relatively little is known regarding its expression pattern and clinical significance in GC. Our results showed that RGMB-AS1 is highly expressed in GC tissue samples and cell lines. Higher RGMB-AS1 expression significantly correlated with tumor size, TNM stage, and lymph node metastasis among the patients with GC. Furthermore, patients with GC in the high-RGMB-AS1 expression group demonstrated shorter overall survival than that of patients in the low-RGMB-AS1 expression group.
In terms of function, RGMB-AS1 is reported to play oncogenic roles during cancer initiation and progression. For example, RGMB-AS1 knockdown inhibits laryngeal squamous cell carcinoma cell proliferation and invasion in vitro and suppresses tumor growth of these cells in vivo.18 In lung cancer, the knockdown of RGMB-AS1 restricts cell growth and metastasis, induces cell cycle arrest at the G1–G0 transition in vitro, and slows tumor growth in vivo.19 In papillary thyroid carcinoma, RGMB-AS1 knockdown attenuates cell proliferation, migration, and invasion in vitro.20 In contrast, RGMB-AS1 serves as a tumor-suppressive lncRNA in hepatocellular carcinoma by inhibiting cell proliferation, migration, and invasion and facilitating apoptosis.21 In the present study, we aimed to test whether a change in RGMB-AS1 expression is implicated in the malignancy of GC in vitro and in vivo. We demonstrated that RGMB-AS1 performs oncogenic functions in the malignancy of GC by inducing cell proliferation, migration, and invasion and by decreasing apoptosis in vitro. Our experiments also revealed that silencing RGMB-AS1 retarded tumor growth of GC cells in vivo.
Mechanistically, the most widely studied latent mechanism of action of lncRNAs is based on the role of a ceRNA that sponges miRNAs to alleviate the repression of their target genes.30–32 Following the identification of the expression profile and functions of RGMB-AS1 in GC, it was necessary to elucidate the mechanisms responsible for the oncogenic activities of RGMB-AS1 in GC progression. Accordingly, we demonstrated that RGMB-AS1, which harbors miR-574–binding sites, can act as an effective sponge toward miR-574, thereby positively modulating HDAC4 expression. MiR-574 is underexpressed in GC, and this underexpression strongly correlates with tumor stage and differentiation status.22 MiR-574 exerts tumor-suppressive actions in GC cells and participates in the control over cell growth, metastasis, epithelial–mesenchymal transition, and cisplatin resistance.22,23 HDAC4, a member of the HDAC family, is overexpressed in GC, and plays multiple roles in the malignant characteristics of GC cells in vitro and in vivo.33–35 Understanding the newly identified RGMB-AS1–miR-574–HDAC4 regulatory network can clarify the oncogenic activities of RGMB-AS1 in GC and may point to promising therapeutic regimens for patients with GC.
Conclusion
In summary, the results of this study suggested that upregulation of RGMB-AS1 in GC correlates with worse clinical outcomes. RGMB-AS1 promotes the aggressive behavior of GC cells in vitro and in vivo, with these oncogenic activities being mediated by miR-574 sponging, thereby increasing HDAC4 expression. Our results thus offer an attractive therapeutic target in GC.
Ethics Approval and Consent to Participate
The study protocol was approved by the Ethics Committee of The Second Hospital of Jilin University. All the participants signed an informed consent form. Animal maintenance and experimental steps were approved by the Animal Care and Use Committee of The Second Hospital of Jilin University. All experimental steps were performed in compliance with the Animal Protection Law of the People’s Republic of China-2009 for experimental animals.
Disclosure
The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Li G, Hu Y, Liu H. Current status of randomized controlled trials for laparoscopic gastric surgery for gastric cancer in China. Asian J Endosc Surg. 2015;8(3):263–267. doi:10.1111/ases.12198
4. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi:10.1016/S0140-6736(16)30354-3
5. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
6. Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi:10.1016/j.cell.2011.03.014
7. Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi:10.1016/j.tcb.2017.11.008
8. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi:10.1016/j.cell.2013.02.012
9. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9). doi:10.3390/cells8091015
10. Cossu AM, Mosca L, Zappavigna S, et al. Long non-coding RNAs as important biomarkers in laryngeal cancer and other head and neck tumours. Int J Mol Sci. 2019;20(14):3444. doi:10.3390/ijms20143444
11. Zhou R, Wu Z, Deng X, Chen H. The long non-coding RNA OLC8 enhances gastric cancer by interaction with IL-11. J Clin Lab Anal. 2019;33:e22962.
12. Deng W, Zhang Y, Cai J, et al. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp Biol Med. 2019;244(12):953–959. doi:10.1177/1535370219860207
13. Liu Y, Zhang YM, Ma FB, Pan SR, Liu BZ. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J Gastroenterol. 2019;25(22):2763–2775. doi:10.3748/wjg.v25.i22.2763
14. Li Y, Zhu G, Ma Y, Qu H. lncRNA CCAT1 contributes to the growth and invasion of gastric cancer via targeting miR-219-1. J Cell Biochem. 2019. doi:10.1002/jcb.26560
15. Zong W, Feng W, Jiang Y, et al. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer. 2019. doi:10.1007/s10120-019-00998-w
16. Pang W, Zhai M, Wang Y, Li Z. Long noncoding RNA SNHG16 silencing inhibits the aggressiveness of gastric cancer via upregulation of microRNA-628-3p and consequent decrease of NRP1. Cancer Manag Res. 2019;11:7263–7277. doi:10.2147/CMAR.S211856
17. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi:10.1016/j.devcel.2006.09.009
18. Xu Z, Xi K. LncRNA RGMB-AS1 promotes laryngeal squamous cell carcinoma cells progression via sponging miR-22/NLRP3 axis. Biomed Pharmacother. 2019;118:109222. doi:10.1016/j.biopha.2019.109222
19. Li P, Zhang G, Li J, et al. Long noncoding RNA RGMB-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS One. 2016;11(3):e0150790. doi:10.1371/journal.pone.0150790
20. Zhang Z, Li SY, Zhang LB. LncRNA RGMB-AS1 is activated by E2F1 and promotes cell proliferation and invasion in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(7):1979–1986. doi:10.26355/eurrev_201804_14725
21. Sheng N, Li Y, Qian R, Li Y. The clinical significance and biological function of lncRNA RGMB-AS1 in hepatocellular carcinoma. Biomed Pharmacother. 2018;98:577–584. doi:10.1016/j.biopha.2017.12.067
22. Su Y, Ni Z, Wang G, et al. Aberrant expression of microRNAs in gastric cancer and biological significance of miR-574-3p. Int Immunopharmacol. 2012;13(4):468–475. doi:10.1016/j.intimp.2012.05.016
23. Wang M, Zhang R, Zhang S, Xu R, Yang Q. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene. 2019;700:110–119. doi:10.1016/j.gene.2019.03.043
24. Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10(10):694. doi:10.1038/s41419-019-1940-3
25. Liu Y, Yin L, Chen C, Zhang X, Wang S. Long non-coding RNA GAS5 inhibits migration and invasion in gastric cancer via interacting with p53 protein. Digest Liver Dis. 2019. doi:10.1016/j.dld.2019.08.012
26. Xiao J, Lai H, Wei SH, Ye ZS, Gong FS, Chen LC. lncRNA HOTAIR promotes gastric cancer proliferation and metastasis via targeting miR-126 to active CXCR4 and RhoA signaling pathway. Cancer Med. 2019. doi:10.1002/cam4.1302
27. Cao C, Xu Y, Du K, et al. LINC01303 functions as a competing endogenous RNA to regulate EZH2 expression by sponging miR-101-3p in gastric cancer. J Cell Mol Med. 2019. doi:10.1111/jcmm.14593
28. Da J, Liu P, Wang R, Bu L. Upregulation of the long non-coding RNA FAM83H-AS1 in gastric cancer and its clinical significance. Pathol Res Pract. 2019;215(10):152616. doi:10.1016/j.prp.2019.152616
29. Li L, Kou J, Zhong B. Up-regulation of long non-coding RNA AWPPH inhibits proliferation and invasion of gastric cancer cells via miR-203a/DKK2 axis. Hum Cell. 2019;32(4):495–503. doi:10.1007/s13577-019-00277-x
30. Zhao Q, Wu C, Wang J, et al. LncRNA SNHG3 promotes hepatocellular tumorigenesis by targeting miR-326. Tohoku J Exp Med. 2019;249(1):43–56. doi:10.1620/tjem.249.43
31. Zhao H, Hu GM, Wang WL, Wang ZH, Fang Y, Liu YL. LncRNA TDRG1 functions as an oncogene in cervical cancer through sponging miR-330-5p to modulate ELK1 expression. Eur Rev Med Pharmacol Sci. 2019;23(17):7295–7306. doi:10.26355/eurrev_201909_18834
32. Gou L, Zou H, Li B. Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther. 2019;20:1–11.
33. Kang ZH, Wang CY, Zhang WL, et al. Histone deacetylase HDAC4 promotes gastric cancer SGC-7901 cells progression via p21 repression. PLoS One. 2014;9(6):e98894. doi:10.1371/journal.pone.0098894
34. Colarossi L, Memeo L, Colarossi C, et al. Inhibition of histone deacetylase 4 increases cytotoxicity of docetaxel in gastric cancer cells. Proteom Clin Appl. 2014;8(11–12):924–931. doi:10.1002/prca.201400058
35. Li Y, Wang K, Wei Y, et al. lncRNA-MIAT regulates cell biological behaviors in gastric cancer through a mechanism involving the miR-29a-3p/HDAC4 axis. Oncol Rep. 2017;38(6):3465–3472. doi:10.3892/or.2017.6020
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.