Back to Journals » OncoTargets and Therapy » Volume 14
Long Noncoding RNA LINC01410 Suppresses Tumorigenesis and Enhances Radiosensitivity in Neuroblastoma Cells Through Regulating miR-545-3p/HK2 Axis
Authors Mou L, Wang L, Zhang S, Wang Q
Received 18 December 2020
Accepted for publication 4 April 2021
Published 18 May 2021 Volume 2021:14 Pages 3225—3238
DOI https://doi.org/10.2147/OTT.S297969
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Liping Mou,1 Lili Wang,2 Shaoming Zhang,3 Qinghua Wang4
1Department of Child Healthcare, People’s Hospital of Rizhao, Rizhao, 276800, Shandong, People’s Republic of China; 2Department of Pediatrics, People’s Hospital of Rizhao, Rizhao, 276800, Shandong, People’s Republic of China; 3Department of Neonatology, People’s Hospital of Rizhao, Rizhao, 276800, Shandong, People’s Republic of China; 4Department of Laboratory, People’s Hospital of Rizhao, Rizhao, 276800, Shandong, People’s Republic of China
Correspondence: Qinghua Wang
Department of Laboratory, People’s Hospital of Rizhao, No. 126, Tai’an Road, Rizhao, 276800, Shandong, People’s Republic of China
Tel +86-633-3365100
Email [email protected]
Background: Abnormal expression of long noncoding RNAs (lncRNAs) was often involved in tumorigenesis and radiosensitivity of various cancers. The aim of this study was to explore the biological function and regulatory mechanism of lncRNA long intergenic non-protein coding RNA 1410 (LINC01410) in tumorigenesis and radiosensitivity of neuroblastoma (NB).
Methods: The expression of LINC01410, microRNA-329-3p (miR-545-3p) and hexokinase 2 (HK2) was detected by quantitative real-time polymerase chain reaction (qRT-PCR). Methylthiazolyldiphenyl tetrazolium bromide (MTT) assay, colony formation assay and transwell assay were utilized to detect cell viability, colony formation and cell invasion abilities. Glucose consumption or lactate production was measured by glucose assay kit or lactate assay kit, respectively. The interaction between miR-545-3p and LINC01410 or HK2 was predicted by starBase v2.0 and verified by dual-luciferase reporter, RNA Immunoprecipitation (RIP) and RNA pull-down assays. Western blot was used to measure the protein expression of HK2. The mice xenograft model was established to investigate the role of LINC01410 in vivo.
Results: LINC01410 and HK2 were highly expressed while miR-545-3p was lowly expressed in NB tissues and cells. LINC01410 knockdown inhibited tumorigenesis by repressing cell proliferation and invasion, and increased the radiosensitivity via inhibiting colony formation rates and glycolysis. LINC01410 knockdown also suppressed tumor growth in vivo. Moreover, miR-545-3p could bind to LINC01410 and its downregulation reversed the effects of LINC01410 knockdown on tumorigenesis and radiosensitivity. Additionally, HK2 was a direct target of miR-545-3p and its overexpression attenuated the effects of miR-545-3p restoration on suppression of tumorigenesis and promotion of radiosensitivity. Besides, LINC01410 functioned as a molecular sponge of miR-545-3p to regulate HK2 expression.
Conclusion: LINC01410 interference inhibited tumorigenesis and increased radiosensitivity via regulating miR-545-3p/HK2 axis, providing a novel therapeutic strategy for NB.
Keywords: NB, LINC01410, miR-545-3p, HK2, tumorigenesis, radiosensitivity
Introduction
Neuroblastoma (NB) is one of the most frequent extracranial childhood tumors and originates from primitive neural crest cells of the sympathetic nervous system, accounting for 7% of malignant tumors in children and approximately 15% of all childhood cancer deaths.1,2 In spite of advances in treatment have improved the survival times of NB patients, children with regional or distant metastatic disease still have a poor prognosis.3 Radiotherapy is the main treatment for tumors, but the efficacy of radiotherapy in treating NB is limited due to obtaining radioresistance in the treatment of this disease.4 Thus, it is necessary to explain the molecular mechanisms of NB and identify more effective therapeutic targets to increase the radiosensitivity.
Long non-coding RNAs (lncRNAs), highly conserved transcripts (>200 nucleotides), lack protein-coding capacity and play regulatory roles in various physiopathology processes.5 Currently, numerous studies have demonstrated that lncRNAs are abnormally expressed in a variety of cancers, including NB.6 For instance, lncRNA NBAT-1 downregulation promoted aggressive NB through enhancing proliferation and attenuating differentiation of neuronal precursors.7 Moreover, lncRNA RMRP was overexpressed in NB tissues and its knockdown suppressed the progression of NB.8 Importantly, recent researches have proven that lncRNA long intergenic non-protein coding RNA 1410 (LINC01410) expression is tightly linked to the progression of multiple cancers, such as thyroid cancer,9 cervical cancer,10 and gastric cancer.11 Besides, pervious study uncovered that LINC01410 was highly expressed in NB tissues and its expression was related to NB development.12 However, the underlying mechanisms of LINC01410 involved in regulating the progression of NB and radiosensitivity remain largely unknown.
Increasing evidence reveals that lncRNAs serve as competing endogenous RNAs (ceRNAs) to modulate gene expression through sponging microRNA (miRNA).13 MiRNAs are usually small RNAs (18–25 nucleotides) that lack protein-coding potential and negatively gene expression via combining with complementary sequences on target mRNA.14,15 Currently, accumulating evidence demonstrated that aberrant expression miRNA was closely related to the development and progression of cancers, and could affect the cells response to radiation.16,17 MiR-545-3p has been suggested to function as a tumor suppressor or tumor promoter in different cancers.18,19 Moreover, miR-545 was also reported to be lowly expressed in NB.20 Nevertheless, the functions of miR-545-3p on the tumorigenesis and radiosensitivity of NB are poorly understood.
Several miRNAs target hexokinase 2 (HK2; a metabolism-related factor) to influence the progression of diverse types of cancer.21,22 HK2 has been reported to be upregulated in NB and acts as a target of miR-143-3p in NB.23 However, there is no report on the interaction between miR-545-3p and HK2, and exact roles of HK2 in NB progression and radiosensitivity should be explored.
In the present study, the expression of LINC01410, miR-545-3p and HK2 was analyzed in NB tissues and cells. In addition, we explored their effects on proliferation, invasion, glycolysis, and radiosensitivity, and investigated the regulatory network of LINC01410/miR-545-3p/HK2 in NB cells, aiming to offer a new biological target for treatment of NB.
Materials and Methods
Clinical Tissue Samples
In this study, thirty NB tissues and paired normal tissues were obtained from patients undergoing surgery at People’s Hospital of Rizhao. The patients did not receive any treatment before surgery. Each patient has signed written informed consent. This research was permitted by Research Ethics Committee of People’s Hospital of Rizhao and performed in accordance with the Declaration of Helsinki. Excised tissues were collected and promptly frozen in liquid nitrogen, and then cryopreserved at −80°C for subsequent study.
Cell Culture and Transfection
Human embryonic kidney (HEK293) and NB cell line (SK-N-BE(2)) were bought from BeNa Culture Collection (Beijing, China). The NB cell line (GI-LI-N) was obtained from Conservation Genetics CAS Kunming Cell Bank (Yunnan, China). These cells were cultivated in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin. The cells were grown in an incubator of CO2 (5%) at 37°C.
For this study, small interfering RNA against LINC01410 (si-LINC01410) and the corresponding control (si-NC), miR-545-3p mimic (miR-545-3p) and the corresponding control (miR-NC), miR-545-3p inhibitor (anti-miR-545-3p) and the corresponding control (anti-miR-NC), LINC01410 or HK2 overexpression plasmid (LINC01410 or HK2) and the corresponding control (pcDNA) were bought from GenePharma (Shanghai, China). Lentivirus-mediated shRNA interference targeting LINC01410 (sh-LINC01410) and the corresponding control (sh-NC) were constructed by Genechem (Shanghai, China). GI-LI-N and SK-N-BE(2) cells were transfected with oligonucleotide (50 nM) or plasmid (2 μg) using Lipofectamine 3000 (Invitrogen).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was applied to isolate total RNA from tissues (NB tissues and normal tissues) and cells (HEK293, GI-LI-N and SK-N-BE(2)). Next, the first strand of complementary DNA (cDNA) was synthesized with a High-Capacity cDNA Reverse Transcription Kit and TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). The qRT-PCR was carried out using the SYBR Green PCR kit (Thermo Fisher Scientific) on the ABI 7300 system (Thermo Fisher Scientific). In this study, the primers were as follows: LINC01410 (Forward, 5ʹ-GTGACAAGAATGGCCCAAGC-3ʹ; Reverse, 5ʹ-ACTGTGCACCTGTTACACCA-3ʹ); miR-545-3p (Forward, 5ʹ-GCCGAGTCAGCAAACATTTAT-3ʹ; Reverse, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); HK2 (Forward, 5ʹ-ACAGCCTGGACGAGAGCATC-3ʹ; Reverse, 5ʹ-AGGTCAAACTCCTCTCGCCG-3ʹ); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Forward, 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ; Reverse, 5ʹ-ATCCGTTGACTCCGACCTTCAC-3ʹ), U6 snRNA (Forward, 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ; Reverse, 5ʹ-ACGCTTCACGAATTTGCGTGTC-3ʹ). The LINC01410, HK2 or miR-545-3p expression was evaluated with the 2−ΔΔCt method, followed by normalizing to GAPDH or U6 snRNA, respectively.
Cell Viability Assay
Methylthiazolyldiphenyl tetrazolium bromide (MTT) was utilized for detecting cell viability. In brief, GI-LI-N and SK-N-BE(2) cell suspension (100 uL, 2×103 cells) were seeded in 96-well plates and then transfected with si-LINC01410, si-LINC01410 + anti-miR-545-3p, miR-545-3p, miR-545-3p + HK2, or matched controls. MTT reagent (5mg/mL, 10 μL, Beyotime, Shanghai, China) was added to each well after transfection. After incubation for 4 h, the cultured medium from per well would be carefully discarded and dimethyl sulfoxide (150 μL) was added. Lastly, the absorbance was evaluated at 490 nm under a microplate reader (Bio-Rad, Hercules, CA, USA).
Irradiation (IR) and Colony Formation Assay
After transfection for 48 h, GI-LI-N and SK-N-BE(2) cells were placed into six-well plates and the medium was updated every three days. For treatment of IR, cells were exposed to different doses of radiation by a linear accelerator (Varian, Palo Alto, CA, USA) at a dose rate of 3.5 Gy/min. After incubation for 2 weeks, GI-LI-N and SK-N-BE(2) cells were carefully washed with cold phosphate-buffered saline (PBS; pH=7.2) and subsequently fixed with paraformaldehyde (4%) for 30 min at 4°C. After that, GI-LI-N and SK-N-BE(2) cells were washed by PBS and then stained by crystal violet (0.1%, Sigma-Aldrich, St. Louis, MO, USA). Microscope (Olympus, Tokyo, Japan) was applied to count colonies containing more than 50 cells. The survival fraction was calculated following the equation: the number of colonies/(number of plated cells × plating efficiency).24
Transwell Assay
Transwell assay was performed using a 24-well plate inserts with 8 μm pores (Corning Incorporated, Corning, NY, USA) to evaluate cell invasion capacity. GI-LI-N and SK-N-BE(2) cells (2×104 cells/well) re-suspended in DMEM medium alone (100 μL) were placed into the top chamber pre-coated with Matrigel (BD Bioscience, Franklin Lakes, NJ, USA). To induce cells invading through the membrane, the bottom chamber was filled with DMEM containing 10% FBS. After 24 h of incubation, non-invaded cells were carefully removed with cotton bud, and invaded cells were fixed using ethanol (95%) and then stained using crystal violet (0.1%). Finally, a microscope (Olympus, Tokyo, Japan) was applied to observe and count invaded cells in five random fields.
Measurement of Glucose Consumption and Lactate Production
In accordance with the manufacturer’s instructions, the glucose consumption and lactate production were detected with the glucose assay kit (Sigma-Aldrich) and lactate assay kit (BioVision, Mountain View, CA, USA), respectively. A standard calibration curve was used for determining the data, followed by normalizing to the amount of total protein. The consumption glucose and lactate production were analyzed by detecting the absorbance of each tube against the reagent blank at 540 nm and 570 nm, respectively.
Dual-Luciferase Reporter Assay
The possible binding sites of miR-545-3p and LINC01410 or HK2 were predicted by starBase v2.0. The LINC01410 and HK2 3ʹUTR fragments that contained wild-type (WT) or mutant (MUT) binding sites of miR-545-3p were amplified and inserted into pmirGlO luciferase reporter vector (Promega, Madison, WI, USA) to create the WT plasmids (WT-LINC01410, HK2 3ʹUTR-WT) or MUT plasmids (MUT-LINC01410, HK2 3ʹUTR-MUT). Afterward, GI-LI-N and SK-N-BE(2) cells were co-transfected with reporter plasmids and miR-545-3p or miR-NC. After incubating the cells for 48 h, the luciferase activity was estimated via Dual-Luciferase Reporter assay system (Promega).
RNA Immunoprecipitation (RIP) Assay
EZ-Magna RIP Kit (Millipore, Billerica, MA, USA) was used to validate the interaction between miR-545-3p and LINC01410 or HK2. In brief, cells were lysed by complete RIP lysis buffer. After that, whole cell extract (100 µL) was incubated by RIP buffer containing magnetic beads coated with human anti-argonaute2 (Ago2) or anti-immunoglobulin G (IgG) (as control). Next, samples were digested using the proteinase K. Finally, the RNAs in the immunoprecipitates were isolated with Trizol reagent, and the expression levels of LINC01410, miR-545-3p and HK2 were examined by qRT-PCR.
RNA Pull-Down Assay
RNA pull-down assay was performed to verify the endogenous interaction between miR-545-3p and LINC01410 or HK2 in NB cells. Biotinylated miRNAs (bio-miR-NC and bio-miR-545-3p) were obtained from Sangon Biotech (Shanghai, China). In brief, cells transfected with bio-miR-NC or bio- miR-545-3p were lysed by lysis buffer. The cell lysates were incubated with Dynabeads M-280 Streptavidin (Thermo Fisher Scientific). Next, the beads were washed, the RNA complexes bound to the beads were extracted with Trizol reagent. Finally, the enrichment of LINC01410 and HK2 was detected by qRT-PCR.
Western Blot Assay
Transfected cells were lysed using RIPA lysis buffer (Sigma-Aldrich) supplemented with 1mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich) to extract the total protein. After quantification by using bicinchoninic acid protein assay kit (Tanon, Shanghai, China), 30 μg of protein in each group was separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After that, polyvinylidene fluoride (Beyotime) membranes were employed to transfer the protein. After blocking with 5% non-fat milk in PBS containing 0.05% Tween 20 (PBST), the membranes were immunoblotted by primary antibody against HK2 (1:5000, ab227198, Abcam, Cambridge, UK) or GAPDH (1:2500, ab9485, Abcam) for 12–16 h at 4°C. Afterward, membranes were maintained in horseradish peroxidase-conjugated anti-rabbit IgG (1:4000, D110058, Sangon Biotech) after washing with PBST. Immunoreactive bands were visualized through the enhanced chemiluminescence (ECL; Tanon) reagent following the manufacturer’s protocol. ImageJ software was utilized to quantify the protein expression. GAPDH was used as an internal control.
In vivo Tumor Growth Assay
BALB/c nude mice (male, 5-week-old) were purchased from Vital River Laboratory Animal Technology Co., Ltd, (Beijing, China). The sh-LINC01410 or sh-NC was transfected into SK-N-BE(2) cells. Subsequently, stably transfected cells (3×106) were subcutaneously injected in BALB/c nude mice. From the 8th day, tumor volume was examined using a caliper every 4 days and calculated with the formula: length × width2/2. After injection for 10 days, sh-LINC01410 group or sh-NC group were randomly divided into two groups. One group was irradiated with 6Gy X-ray once a week, and the other group served as control. All mice were sacrificed after injection for 4 weeks, and tumor samples were weighted and collected for detection of LINC01410, miR-545-3p and HK2 expression levels. The animal experiments were approved by the Committee on the Ethics of Animal Experiments of People’s Hospital of Rizhao. All animal procedures were conducted following the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health”.
Immunohistochemistry (IHC) Analysis
Tumor tissues from transplanted nude mice were fixed in paraformaldehyde (4%, Beyotime) and then were dehydrated, embedded in paraffin, and cut. After that, consecutive tumor sections (4 μm in thickness) were incubated by antibodies against HK2 (1:200, ab227198, Abcam) overnight at 4°C, followed by incubation with secondary antibody (1:5000, ab205718, Abcam) for 1 h. The sections were stained by diaminobenzidine (DAB), and cell nucleus was counterstained with haematoxylin. At last, stained sections were photographed using a fluorescence microscope (Olympus).
Statistical Analysis
All data in this study were displayed as mean ± standard deviation (SD) and all experiments were repeated at least 3 times. Correlation between miR-545-3p and LINC01410 or HK2 was detected by Spearman rank correlation. The results from different groups were assessed using the two-tailed Student’s t-test (for two groups) and a one-way analysis of variance (ANOVA; for more than two groups). Statistical analyses were performed using GraphPad Prism version 6.0 software (GraphPad Software, San Diego California, USA). P<0.05 indicates a statistically significant result. (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Results
LINC01410 Was Upregulated and miR-545-3p Was Downregulated in NB Tissues and Cells
Firstly, we explored the expression of LINC01410 and miR-545-3p in NB tissues and cells by qRT-PCR. As presented in Figure 1A and B, LINC01410 was overexpressed in NB tissues and cells in contrast to their matched controls. Moreover, we found that the expression of miR-545-3p was decreased in NB tissues and cells compared to their corresponding controls (Figure 1C and D). Besides, the correlation between of LINC01410 and miR-545-3p was analyzed in NB tissues. As depicted in Figure 1E, LINC01410 expression was negatively correlated with miR-545-3p in NB tissues (r=−0.5852, p<0.001). These data indicated that LINC01410 and miR-545-3p might be involved in the pathogenesis of NB.
LINC01410 Knockdown Inhibited Cell Proliferation and Invasion and Enhanced Radiosensitivity by Inhibiting Glycolysis in NB Cells
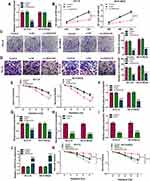
Next, the effects of LINC01410 on cell proliferation, invasion, glycolysis, and radiosensitivity were investigated using loss-function experiments. The transfection efficiency of si-LINC01410 was determined by qRT-PCR. The results showed that the expression of LINC01410 was decreased in GI-LI-N and SK-N-BE(2) cells transfected with si-LINC01410 compared to those cells transfected with si-NC or control cells (Figure 2A), suggesting the transfection of si-LINC01410 was successful. MTT assay revealed that LINC01410 interference decreased GI-LI-N and SK-N-BE(2) cell viability (Figure 2B). Moreover, colony formation assay indicated that LINC01410 deficiency decreased the number of colonies in GI-LI-N and SK-N-BE(2) cells (Figure 2C). Furthermore, silencing LINC01410 repressed GI-LI-N and SK-N-BE(2) cell invasion ability (Figure 2D). GI-LI-N and SK-N-BE(2) cells were transfected with si-LINC01410 or si-NC and then irradiated with 0 Gy to 8 Gy to explore the effect of LINC01410 on radiosensitivity. The data showed that LINC01410 downregulation strikingly decreased the survival fraction of GI-LI-N and SK-N-BE(2) cells exposed to radiation compared to si-NC or control group (Figure 2E). Most cancer cells mainly rely on aerobic glycolysis to produce the energy needed for cellular processes.25 This aerobic glycolysis will result in increased glucose consumption and lactate production. We observed that knockdown of LINC01410 inhibited glucose consumption and lactate production in GI-LI-N and SK-N-BE(2) cells (Figure 2F and G). Moreover, we found that glucose consumption and lactate production were decreased in GI-LI-N and SK-N-BE(2) cells treated with 2-deoxyglucose (2-DG; glycolytic inhibitor) (Figure 2H and I). QRT-PCR analysis showed that the expression of LINC01410 was enhanced in cells transfected with LINC01410 compared to pcDNA group (Figure 2J). Besides, the promotive effect of LINC01410 on cell survival fraction was overturned by treatment with 2-DG (Figure 2K), suggesting that LINC01410 knockdown enhanced the radiosensitivity through inhibiting glycolysis. Taken together, these results demonstrated that LINC01410 interference inhibited the tumorigenesis and increased the radiosensitivity in NB cells.
LINC01410 Directly Interacted with miR-545-3p in NB Cells
In view of the alteration of LINC01410 and miR-545-3p in NB tissues and cell lines and the negative correction between LINC01410 and miR-545-3p expression, we wondered whether the function of LINC01410 was mediated by miR-545-3p through complementary binding sites. StarBase v2.0 predicted that LINC01410 contained putative binding sites of miR-545-3p (Figure 3A). Transfection of miR-545-3p increased the expression of miR-545-3p expression, while transfection of anti-miR-545-3p decreased miR-545-3p expression in GI-LI-N and SK-N-BE(2) cells (Figure 3B). To verify whether miR-545-3p was a direct target of LINC01410, the dual-luciferase reporter assay, RIP assay and RNA pull-down assay were performed. As presented in Figure 3C, overexpression of miR-545-3p led to a significant decrease in the luciferase activity of WT-LINC01410, while luciferase activity of MUT-LINC01410 was not evidently affected after transfection with miR-545-3p in GI-LI-N and SK-N-BE(2) cells. RIP assay revealed that both miR-545-3p and LINC01410 were immunoprecipitated in argonaute2 (Ago2) group instead of immunoglobulin G (IgG) group (Figure 3D). RNA pull-down assay showed that the expression of LINC01410 was significantly elevated in biotinylated miR-545-3p (bio-miR-545-3p) group rather than biotinylated negative (bio-miR-NC) group (Figure 3E), suggesting that miR-545-3p could pull down LINC01410. Moreover, the expression of LINC01410 was reduced in GI-LI-N and SK-N-BE(2) cells transfected with si-LINC01410, while its expression was elevated in cells transfected with LINC01410 (Figure 3F). Next, we explored the effect of LINC01410 on expression of miR-545-3p. As expected, knockdown of LINC01410 increased the expression of miR-545-3p, whereas overexpression of LINC01410 presented an opposite effect (Figure 3G), suggesting that miR-545-3p was negatively regulated by LINC01410. Meanwhile, we observed that co-transfection of anti-miR-545-3p and si-LINC01410 abated the effect of si-LINC01410 on promotion of miR-545-3p expression (Figure 3H). Additionally, inhibition of miR-545-3p reversed the inhibitory effects of LINC01410 knockdown on viability, colony formation and invasion of GI-LI-N and SK-N-BE(2) cells (Figure 3I–K). Moreover, the suppressive effect of LINC01410 interference on the survival fraction was abolished by downregulation of miR-545-3p in GI-LI-N and SK-N-BE(2) cells exposed to IR (Figure 3L). Furthermore, miR-545-3p silence abated the inhibitory effect of LINC01410 knockdown on glucose consumption and lactate production (Figure 3M and N). Our findings suggested that miR-545-3p could bind to LINC01410 and its knockdown reversed the effects of LINC01410 interference on tumorigenesis and radiosensitivity in NB cells.
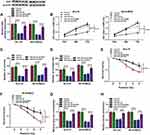
HK2 Was a Direct Target of miR-545-3p in NB Cells
To explore the underlying mechanism of miR-545-3p in progression of NB, the function targets of miR-545-3p were searched by starBase v2.0. As presented in Figure 4A, miR-545-3p and HK2 3ʹUTR had complementary binding sequence, indicating that HK2 might be a target of miR-545-3p. Dual-luciferase reporter assay displayed that the luciferase activity of HK2 3ʹUTR-WT was obviously inhibited by transfection of miR-545-3p, whereas no clear change of the luciferase activity of HK2 3ʹUTR-MUT was found (Figure 4B). RIP assay demonstrated that miR-545-3p and HK2 were obviously enriched in the Ago2 immunoprecipitation compared with the IgG immunoprecipitation (Figure 4C). Moreover, HK2 was strikingly enriched in GI-LI-N and SK-N-BE(2) cells transfected with bio-miR-545-3p (Figure 4D). Next, we detected the expression of HK2 in NB tissues and cells. The results suggested that the mRNA expression and protein expression of HK2 were increased in NB tissues in comparison with normal tissues (Figure 4E and F). Additionally, HK2 expression was negatively correlated with miR-545-3p level in NB tissues (r=−0.5689, p<0.001) (Figure 4G). Likewise, the mRNA expression and protein expression of HK2 were also enhanced in NB cells (GI-LI-N and SK-N-BE(2)) compared to HEK293 cells (Figure 4H). These data illustrated that miR-545-3p directly interacted with HK2.
Overexpression of HK2 Reversed the Effects of miR-545-3p on Inhibition of Tumorigenesis and Promotion of Radiosensitivity in NB Cells
To determine whether the effects of miR-545-3p were regulated by HK2 expression, GI-LI-N and SK-N-BE(2) cells were transfected with miR-NC, miR-545-3p, miR-545-3p + pcDNA, or miR-545-3p + HK2. Western blot showed that HK2 protein expression was decreased in cells transfected with miR-545-3p, while the effect was abated by co-transfection of HK2 (Figure 5A). Additionally, overexpression of miR-545-3p reduced the cell viability, colony formation and invasion of GI-LI-N and SK-N-BE(2) cells, which was reversed by upregulating HK2 (Figure 5B–D). Moreover, the inhibitory effects of miR-545-3p upregulation on survival fraction and glycolysis were also overturned by overexpression of HK2 (Figure 5E–H). Collectively, these findings demonstrated that miR-545-3p exerted its biological functions through regulating HK2 expression.
HK2 Was Regulated by LINC01410 and miR-545-3p in NB Cells
Next, knockdown efficiency of miR-545-3p was tested by qRT-PCR. As shown in Figure 6A, the expression of miR-545-3p was significantly reduced in GI-LI-N and SK-N-BE(2) cells transfected with anti-miR-545-3p compared to those cells transfected with anti-miR-NC or control group. To explore whether LINC01410 served as a ceRNA of miR-545-3p to modulate HK2 expression, GI-LI-N and SK-N-BE(2) cells were transfected with si-NC, si-LINC01410, si-LINC01410 + anti-miR-NC, or si-LINC01410 + anti-miR-545-3p. Western blot assay demonstrated that the protein level of HK2 was decreased after transfection with si-LINC01410, while the effect was reversed by downregulating miR-545-3p (Figure 6B). In a word, our results proved that LINC01410 regulated HK2 expression by sponging miR-545-3p in NB cells.
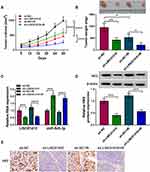
Downregulation of LINC01410 Inhibited Tumor Growth and Increased Radiosensitivity by Regulating miR-545-3p and HK2 in vivo
To explore the effect of LINC01410 on tumor growth and radiation response in vivo, we established a xenograft model in which the SK-N-BE(2) cells stably transfected with sh-LINC01410 or sh-NC were subcutaneously injected into BALB/c nude mice and irradiated with 6Gy X-ray once a week or did not irradiate. In line with in vitro results, LINC01410 knockdown or IR treatment suppressed tumor volume and weight, and combination of LINC01410 interference and IR treatment significantly inhibited tumor volume and weight in xenograft model compared with only sh-LINC01410 group or IR group (Figure 7A and B). In addition, knockdown of LINC01410 decreased the expression of LINC01410 and HK2 while increased the expression of miR-545-3p in excised tumor masses (Figure 7C and D). IHC analysis showed that LINC01410 deficiency reduced HK2-positive cells (Figure 7E), suggesting that LINC01410 knockdown decreased the expression HK2 in tumor tissues. These above findings indicated that downregulation of LINC01410 could enhance the radiosensitivity to inhibit tumor growth in vivo.
Discussion
NB is considered to be a common and aggressive malignancy in children. Recently, it has been increasingly recognized that dysregulation of lncRNAs is involved in the progression of NB.23,26 Radiotherapy has become an effective strategy for NB treatment, however, the influence of lncRNAs on radiosensitivity of NB cells is not well elucidated. The purpose of this research was to explore the effects of LINC01410 on NB progression and radiosensitivity.
Up to now, a growing number of researches have shown that LINC01410 serves as an oncogene and plays essential roles in cellular behaviors, such as cell growth, cell cycle, metastasis and apoptosis.27 Besides, Lu et al pointed out that LINC01410 was overexpressed in endometrial cancer tissues, and accelerated endometrial cancer cell progression through miR-23c/CHD7 axis.28 Moreover, Jiang et al uncovered that LINC01410 knockdown inhibited cholangiocarcinoma cell proliferation and metastasis via sponging miR-124-3p to decrease SMAD5 expression.29 In our research, it was found that LINC01410 abundance was elevated in NB tissues and cells. Additionally, interference of LINC01410 suppressed cell proliferation and invasion, and elevated radiosensitivity via suppressing glycolysis in NB cells as well as restrained tumor growth through enhancing radiosensitivity in vivo. In agreement with our findings, Mi et al disclosed that LINC01410 expression was markedly enhanced in NB tissues and its knockdown inhibited the cell growth and tumor growth and facilitated the apoptosis in NB cells by regulating miR-506-3p/WEE1 axis.12 These results revealed that LINC01410 was involved in the tumorigenesis of NB and radiosensitivity.
Precious research has shown that miR-545-3p has pivotal roles in the regulating cellular behaviors, such as proliferation, differentiation and migration.30,31 It has recently been identified as a cancer-related miRNA for several cancers. For example, Changjun et al declared that miR-545-3p was highly expressed in hepatocellular carcinoma tissues and cells, and miR-545-3p inhibition limited hepatocellular carcinoma cell proliferation and metastasis via regulating MT1M.18 In contrast, Shi et al proved that miR-545-3p acted as a tumor-suppressive miRNA through targeting HDAC4 to suppress epithelial ovarian cancer proliferation, migration and invasion.32 These findings suggested that miR-545-3p could exert a tumor-suppressive or tumor-promotive function depending on the type of cancer. More importantly, Chen et al showed that miR-545 was a downregulated miRNA in NB,20 suggesting that miR-545-3p might act as a tumor suppressor in NB. Here, we found that miR-545-3p abundance was declined in NB tissues and cells, and was inversely correlated with LINC01410. Previous studies have demonstrated that lncRNAs can execute their functions through binding with their downstream miRNAs.33 To further explore the relationship between LINC01410 and miR-545-3p, starBase v2.0 was used to predict the targeting relationship. Interestingly, starBase v2.0 showed that miR-545-3p contained predicted binding sites with LINC01410. Next, the prediction was validated through dual-luciferase reporter, RIP and RNA pull-down assays. In addition, rescue experiments demonstrated that miR-545-3p knockdown could reverse the impact of LINC01410 interference on cell proliferation, invasion, radiosensitivity, and glycolysis in NB cells. Collectively, our data indicated that LINC01410 exerted its function through regulating miR-545-3p expression.
It is well known that the biological functions of miRNAs are realized via modulating mRNA expression, so the potential target genes for miR-545-3p would be analyzed in further analysis. Despite the fact that numerous tumor-associated genes were predicted using starBase v2.0, HK2 was selected as the candidate target gene of miR-545-3p due to its tumor-promoting effect. Further, we proved that miR-545-3p directly targeted HK2. HK2 (a major type of hexokinase family) was reported to be overexpressed and facilitated rates of glucose metabolism necessary for tumor growth in multiple cancers.34,35 In addition, Botzer et al presented that high expression of HK2 promoted NB metastasis.36 Besides, Cen et al revealed that HK2 was upregulated in NB tissues and cells, and HK2 accumulation weakened the repressive effects of miR-143-3p restoration on progression of NB.23 In line with previous findings, we also demonstrated that HK2 was highly expressed in NB tissues and cells. Moreover, HK2 upregulation could weaken the influence of miR-545-3p restoration on inhibition of tumorigenesis and promotion of radiosensitivity in NB cells. Furthermore, we uncovered that LINC01410 acted as a molecular sponge of miR-545-3p to modulate HK2 expression. In a word, these findings disclosed that LINC01410 regulated the progression and radiosensitivity of NB cells by sponging miR-545-3p to modulate HK2 expression.
In conclusion, LINC01410 and HK2 were upregulated while miR-545-3p was downregulated in NB tissues and cells. Knockdown of LINC01410 inhibited the tumorigenesis and enhanced the radiosensitivity in NB cells by regulating miR-545-3p and HK2 expression. These findings might provide a potential therapeutic strategy for NB.
Acknowledgment
Thanks for all participants involved in this study.
Disclosure
The authors declare that they have no competing interests.
References
1. Matthay KK. Neuroblastoma: biology and therapy. Oncology. 1997;11(12):1857.
2. Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):83–103. doi:10.3322/caac.21219
3. Harrison J, Myers M, Rowen M, Vermund H. Results of combination chemotherapy, surgery, and radiotherapy in children with neuroblastoma. Cancer. 1974;34(3):485–490. doi:10.1002/1097-0142(197409)34:3<485::AID-CNCR2820340302>3.0.CO;2-8
4. Madhusoodhanan R, Natarajan M, Veeraraghavan J, et al. NFκB signaling related molecular alterations in human neuroblastoma cells after fractionated irradiation. J Radiat Res (Tokyo). 2009;50(4):311–324. doi:10.1269/jrr.08110
5. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
6. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
7. Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–737. doi:10.1016/j.ccell.2014.09.014
8. Pan J, Zhang D, Zhang J, Qin P, Wang J. LncRNA RMRP silence curbs neonatal neuroblastoma progression by regulating microRNA-206/tachykinin-1 receptor axis via inactivating extracellular signal-regulated kinases. Cancer Biol Ther. 2019;20(5):653–665. doi:10.1080/15384047.2018.1550568
9. Wang G, Wang X, Jin Y. LINC01410/miR-3619-5p/FOXM1 feedback loop regulates papillary thyroid carcinoma cell proliferation and apoptosis. Cancer Biother Radiopharm. 2019;34(9):572–580. doi:10.1089/cbr.2019.2854
10. Liu F, Wen C. LINC01410 knockdown suppresses cervical cancer growth and invasion via Targeting miR-2467-3p/VOPP1 axis. Cancer Manag Res. 2020;12:855–861. doi:10.2147/CMAR.S236832
11. Zhang JX, Chen ZH, Chen DL, et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37(20):2660–2675. doi:10.1038/s41388-018-0162-y
12. Mi J, Han Y, Zhang J, Hao X, Xing M, Shang C. Long noncoding RNA LINC01410 promotes the tumorigenesis of neuroblastoma cells by sponging microRNA-506-3p and modulating WEE1. Cancer Med. 2020;9(21):8133–8143. doi:10.1002/cam4.3398
13. Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
14. Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161.
15. Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610.
16. Dickey JS, Zemp FJ, Martin OA, Kovalchuk O. The role of miRNA in the direct and indirect effects of ionizing radiation. Radiat Environ Biophys. 2011;50(4):491. doi:10.1007/s00411-011-0386-5
17. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15(1):38. doi:10.1186/s12935-015-0185-1
18. Changjun L, Feizhou H, Dezhen P, Zhao L, Xianhai M. MiR-545-3p/MT1M axis regulates cell proliferation, invasion and migration in hepatocellular carcinoma. Biomed Pharmacother. 2018;108:347–354. doi:10.1016/j.biopha.2018.09.009
19. Cui J, Pan G, He Q, Yin L, Guo R, Bi H. MicroRNA-545 targets ZEB2 to inhibit the development of non-small cell lung cancer by inactivating Wnt/β-catenin pathway. Oncol Lett. 2019;18(3):2931–2938. doi:10.3892/ol.2019.10619
20. Chen B, Hua Z, Qin X, Li Z. Integrated microarray to identify the hub miRNAs and constructed miRNA-mRNA network in neuroblastoma via bioinformatics analysis. Neurochem Res. 2020. doi:10.1007/s11064-020-03155-3
21. Yoshino H, Enokida H, Itesako T, et al. Tumor‐suppressive micro RNA‐143/145 cluster targets hexokinase‐2 in renal cell carcinoma. Cancer Sci. 2013;104(12):1567–1574. doi:10.1111/cas.12280
22. Fang R, Xiao T, Fang Z, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287(27):23227–23235. doi:10.1074/jbc.M112.373084
23. Cen Y, Xu L, Huang S, Zhuang S. miR-143-3p functions as a tumor suppressor by targeting HK2 in neuroblastomas. Int J Clin Exp Med. 2019;12(7):8450–8460.
24. Zhou S, Ye W, Ren J, et al. MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. Am J Cancer Res. 2015;5(1):267.
25. Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134(5):703–707. doi:10.1016/j.cell.2008.08.021
26. Bevilacqua V, Gioia U, Di Carlo V, et al. Identification of linc-NeD125, a novel long non coding RNA that hosts miR-125b-1 and negatively controls proliferation of human neuroblastoma cells. RNA Biol. 2015;12(12):1323–1337. doi:10.1080/15476286.2015.1096488
27. Luo J, Guo Y, Liu X, Yang X, Xiao F, Zhou M. Long non-coding RNA LINC01410 promotes colon cancer cell proliferation and invasion by inhibiting miR-3128. Exp Ther Med. 2018;16(6):4824–4830. doi:10.3892/etm.2018.6806
28. Lu M, Ding N, Zhuang S, Li Y. LINC01410/miR-23c/CHD7 functions as a ceRNA network to affect the prognosis of patients with endometrial cancer and strengthen the malignant properties of endometrial cancer cells. Mol Cell Biochem. 2020;469(1–2):9–19.
29. Jiang T, Wang C, Zhu Y, Han H. LINC01410 promotes cell proliferation and migration of cholangiocarcinoma through modulating miR-124-3p/SMAD5 axis. J Gene Med. 2020;22(6):e3162. doi:10.1002/jgm.3162
30. Hao R, Wang B, Wang H, Huo Y, Lu Y. lncRNA TUG1 promotes proliferation and differentiation of osteoblasts by regulating the miR-545-3p/CNR2 axis. Braz J Med Biol Res. 2020;53(11):e9798. doi:10.1590/1414-431x20209798
31. Zhong Y, Wang Y, Dang H, Wu X. LncRNA AFAP1-AS1 contributes to the progression of endometrial carcinoma by regulating miR-545-3p/VEGFA pathway. Mol Cell Probes. 2020;53:101606. doi:10.1016/j.mcp.2020.101606
32. Shi J, Xu X, Zhang D, et al. Long non-coding RNA PTPRG-AS1 promotes cell tumorigenicity in epithelial ovarian cancer by decoying microRNA-545-3p and consequently enhancing HDAC4 expression. J Ovarian Res. 2020;13(1):127. doi:10.1186/s13048-020-00723-7
33. Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233(9):7164–7172. doi:10.1002/jcp.26543
34. He H-C, Bi X-C, Zheng Z-W, et al. Real-time quantitative RT-PCR assessment of PIM-1 and hK2 mRNA expression in benign prostate hyperplasia and prostate cancer. Med Oncol. 2009;26(3):303–308. doi:10.1007/s12032-008-9120-9
35. Mathupala S, Ko YH, Pedersen PL. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25(34):4777. doi:10.1038/sj.onc.1209603
36. Botzer LE, Maman S, Sagi-Assif O, et al. Hexokinase 2 is a determinant of neuroblastoma metastasis. Br J Cancer. 2016;114(7):759. doi:10.1038/bjc.2016.26
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.