Back to Journals » OncoTargets and Therapy » Volume 12
Long noncoding RNA DLEU1 aggravates glioma progression via the miR-421/MEF2D axis
Authors Feng L, He M, Rao M, Diao J, Zhu Y
Received 4 March 2019
Accepted for publication 5 June 2019
Published 8 July 2019 Volume 2019:12 Pages 5405—5414
DOI https://doi.org/10.2147/OTT.S207542
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
This paper has been retracted.
Li Feng, 1,* Mingyuan He, 1,* Min Rao, 2 Jiandong Diao, 3 Yonggang Zhu 1
1Department of Radiotherapy, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China; 2Department of Gastroenterology, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China; 3Department of Oncology and Hematology, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China
*These authors contributed equally to this work
Background: Long noncoding RNA (lncRNA) deleted in lymphocytic leukemia 1 (DLEU1) was reported to be involved in the development and progression of multiple cancers. However, the accurate expression pattern, biological function and potential molecular mechanism of DLEU1 in glioma are not yet known. The present study investigated the role of DLEU in the development and progression of glioma, as well as the potential mechanism played by DLEU1 in glioma.
Materials and methods: The levels of DLEUI in glioma tissues and cell lines were examined using quantitative real-time PCR. The potential effects of DLEU1 on the proliferation, mobility, invasion and apoptosis of glioma cells were evaluated using corresponding in vitro experiments. The association between DLEU1 and microRNA (miR)-421 was also determined using luciferase reporter activity and RNA immunoprecipitation (RIP) assays.
Results: The results revealed that DLEU1 was significantly upregulated in glioma tissues and cell lines. Increased DLEU1 was positively associated with the high-grade carcinoma (III–IV). Functional studies revealed that knockdown of DLEU1 expression by siRNA led to decreased proliferation, migration and invasion and increased apoptosis in human glioma cells. Furthermore, luciferase reporter activity and RIP assays confirmed that DLEUI could act as a competing endogenous RNA (ceRNA) for miR-421 that functioned as a tumor suppressor in glioma. Moreover, inhibition miR-421 partially restored the effect of DLEU1 knockdown on the glioma cells. DLEU1 could regulate myocyte enhancer factor 2D (MEF2D) expression, a known target of miR-421 in glioma cells.
Conclusion: Taken together, these findings suggested that DLEU1 regulated MEF2D expression to promote glioma progression by sponging miR-421 and that DLEU1 might be a potential therapeutic target for glioma.
Keywords: glioma, LncRNA, DLEU1, miR-421, MEF2D
Introduction
Glioblastoma (GBM) is the most malignant form of glioma with high morbidity and high mortality.1 Despite great progress has been made in GBM treatment, including surgical resection, radio-chemotherapy, or resection combined with postoperative radio-chemotherapy, patients with GBM remain poor prognosis.2,3 Therefore, it is important to understand the pathogenesis of GBM for finding new effective therapeutic targets.
Long noncoding RNAs (lncRNAs), a new member of the noncoding RNA family, were reported to play critical roles in modulating various biological processes, including cellular growth, differentiation, apoptosis and cell cycle progression.4,5 There is increasing evidence demonstrating that abnormal alterations of lncRNAs were involved in cancer initiation, progression, development and metastasis.6,7 Many cancer-specific lncRNAs were found to implicate in glioma progression and serve as potential therapeutic targets for glioma.8,9
The lncRNA deleted in lymphocytic leukemia 1 (DLEU1), located on human chromosomal band 13q14.3, is frequently upregulated, and function as oncogenic lncRNA in gastric cancer,10 ovarian cancer,11 colorectal cancer,12 pancreatic ductal adenocarcinoma,13 endometrial cancer,14 non-small cell lung cancer,15 cervical cancer16 and oral squamous cell carcinoma.17 However, the accurate expression pattern and functional role of DLEU1 in glioma remain unknown. The present study aimed to investigate the expression of DLEU1 level in glioma tissues and cell lines, elucidate the functional role and underlying molecular mechanism of DLEU1 in glioma progression.
Materials and methods
Tissue samples
Forty-two patients with pathologically diagnosed glioma who underwent surgery at the First Hospital of Jilin University between March 2017 and March 2018 were enrolled in this study. None of these subjects received radiotherapy and chemotherapy prior to surgery. In all, 42 paired glioma tissues and matched adjacent brain tissues were immediately obtained from fresh surgical specimens and were then immediately stored in liquid nitrogen. The histological grade of all glioma tissues was classified by two experienced pathologists using 2016 WHO criteria. The detail information of the patients is listed in Table 1. Written informed consent was obtained from all patients and/or their family. This study was in accordance with the Declaration of Helsinki and approved by the Ethical Committee of the China-Japan Union Hospital of Jilin University.
 |
Table 1 Correlation between clinicopathological features and DLEU1 expression in 42 patients with glioma |
Cell culture and transfection
Human astrocyte cell line (NHA) and four glioma cell lines (U87, U251, LN229 and A172) were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). All cell lines were maintained in DMEM (Gibco, NY, USA) containing 10% FBS (Gibco) and antibiotics (100 U/mL penicillin/streptomycin) at 37°C and 5% CO2.
To knockdown DLEU1 expression, three different DLEU1 siRNAs (si-DLEU1#1, si-DLEU1#2 and si-DLEU1#3) were synthesized by Genepharma Ltd. (Shanghai, China). Scrambled siRNA (si-NC) was bought from Genepharma Ltd., and used as negative control. miR-421 mimics, control mimics (miR-NC) and miR-421 inhibitor (miR-421 in) were bought from Genepharma Ltd. U87 cells were seeded into 24-well plates overnight, and then were transfected with siRNA (50 nM), mimics (100 nM) or inhibitor (100 nM) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the guidance of the manufacturer’s instructions. The transfection efficacy was assessed by quantitative real-time-PCR at 24 hrs after transfection.
Quantitative real-time PCR
For the detection of DLEU1 and myocyte enhancer factor 2D (MEF2D) expression, total RNA from tissues and cultured glioma cells was extracted using TRIzol reagent (Invitrogen). Single-stranded cDNA was synthesized using a Reverse Transcription Kit (Takara, Dalian, China), then was quantified using SYBR Select Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) under a model 7900 Fast Real-Time PCR System following the manufacturer’s instructions. The TaqMan MicroRNA assay Kit (Thermo Fisher Scientific, Inc.) was used to examine miR-421 under a model 7900 Fast Real-Time PCR System according to the manufacturer’s instructions. The primer sequences used in this study were described as previously.18–20 The relative expression of miR-421 and DLEU1/MEF2D was normalized to that of U6 or GAPDH using the 2−∆∆Cq method.
Cell viability assay
Transfected cells were seeded in 96-well plates at density of 5×103 cells per well and cultured 24–96 hrs. Cell viability assay was determined at 24, 48 and 72 hrs after incubation using a Cell Counting kit-8 (Dojindo, Kumamoto, Japan) following the manufacturer’s instructions. The absorbance at 450 nm was measured using a Benchmark Plus microplate spectrometer (Bio-Rad Laboratories, Hercules, CA, USA).
Cell apoptosis assay
Cell apoptosis was determined at 48 hrs after transfection using ApoScreen Annexin V Apoptosis Kit (Southern Biotech, Birmingham, AL, USA) on a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturers’ instructions. The apoptosis ratio was analyzed using FlowJo software 3.4 (FlowJo, LLC, Ashland, OR, USA).
Cell migration and invasion assays
Cell invasion and migration were examined using transwell chambers (24-well plate, 8.0 μm; Corning Inc., Corning, NY, USA). Transfected cells resuspended in serum-free DMEM medium were plated into the upper chamber (without Matrigel for migration, with Matrigel for invasion assay), while DMEM medium (600 μL) containing 10% FBS was plated into bottom chamber as a chemoattractant. Forty-eight hours after incubation, migrated or invaded cells on the bottom surface of the chamber were fixed and stained with using a Diff-Quik staining kit (Sysmex, Tokyo, Japan). The number of migrated or invaded cells was counted in five randomly selected microscope under an inverted phase-contrast Microscope (Olympus, Tokyo, Japan).
Dual luciferase assay
The binding sites between DLEU1 and miR-421 were predicted using Starbase2.0 (http://starbase.sysu.edu.cn/). The DLEU1 3ʹUTR sequence containing the theoretical binding sites of miR-421 in DLEU1 and its mutant sequences were synthesized and inserted into the pmirGLO Vector (Promega Corp., Madison, WI, USA), and named as WT-DLEU1 or MT-DLEU1. For the reporter assays, U87 cells were cotransfected with luciferase reporter vector (WT/MT-DLEU1), along with miR-421 mimics or miR-NC using Lipofectamine 3000. At 48 hrs post-transfection, dual luciferase reporter assay system (Promega Corp.) was applied to examine firefly and renilla luciferase activities. The relative luciferase activity in this study was normalized to the Renilla luciferase activity.
RNA immunoprecipitation assay (RIP)
Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was applied to conduct RIP assay according to manufacturer’s protocols. Briefly, cells were harvested and lysed in complete RNA lysis buffer. Then, cell lysate was incubated with human anti-Ago2 antibody (Millipore) and mouse IgG (as a negative control; Millipore). Samples were incubated with Proteinase K buffer (provided by kits) to digest proteins. Subsequently, immunoprecipitated RNA was isolated and subjected to quantitative real-time PCR as above-mentioned.
Western blot analysis
The proteins were extracted and measured from cultured cells as described previously.21 The mouse anti-human MEF2D monoclonal antibody, mouse anti-human GAPDH monoclonal antibody and HRP-conjugated secondary antibodies were bought from Cell Signaling Technology (Danvers, MA, USA). The protein bands were visualized by enhanced chemiluminescence kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
Statistical analysis
All results are expressed as the mean ± SD from at least three independent repeats of the experiments. Student’s t-test was used for comparisons between two groups. One-way ANOVA was used for comparisons of >2 groups. The correlation in a data-set was analyzed using Spearman’s correlation analysis. In all cases, P<0.05 was considered to be a statistically significant.
Results
DLEU1 was upregulated in glioma tissues and cell lines
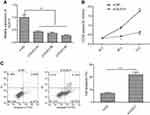
To test the biological roles of DLEU1 in glioma, we first examined the expression of DLEU1 in glioma and adjacent normal tissues by quantitative real-time PCR. As presented in Figure 1A, the median expression of DLEU1 was higher in glioma tissues than that in adjacent normal brain tissues. To explore the clinical significance of DLEU1 in glioma patients, the median expression level of DLEU1 was used as a cutoff point to divide 42 patients into two groups: high expression of DLEU1 (n=24) and low expression of DLEU1 (n=18). Chi-square analysis indicated that high DLEU1 expression was significantly positive associated with advanced WHO grade (P=0.0005) and lower Karnofsky Performance Status (KPS) score (P=0.01). However, no significant difference in DLEU1 expression was observed with age, gender, and tumor size (all P>0.05, Table 1). We also examined the expression of DLEU1 in glioma cell lines and found that the expression of DLEU1 was upregulated in glioma cell lines (U87, U251, LN229 and A172) compared with human astrocyte cell line (NHA) (Figure 1B). These results suggested that DLEU1 play a crucial role in glioma progression.
DLEU1 knockdown inhibits glioma cell proliferation and promotes cell apoptosis
To evaluate the functional role of DLEU1 in glioma, we decreased the expression of DLEU1 in U87 cells using three siRNAs against DLEU1. The results of quantitative real-time PCR demonstrated that all three different DLEU1 siRNAs (si-DLEU1#1, si-DLEU1#2 and si-DLEU1#3) caused a significant reduction in DLEU1 expression (Figure 2A). si-DLEU1#3 displayed the most effect than other siRNAs, thus, it was selected for subsequently all experiments, and referred to as si-DLEU1. The CCK-8 assay revealed that DLEU1 knockdown significantly repressed the proliferation of U87 cells at 48–72 hrs (Figure 2B). Flow cemetery analysis demonstrated that downregulation of DLEU1 significantly promoted cell apoptosis of U87 cells (Figure 2C).
Dleu1knockdown inhibits glioma cell migration and invasion
It was well known that cancer cell metastasis was closely associated with cell migration and invasion.22 To investigate whether DLEUI can affect glioma cell metastasis, we examined the migration and invasion ability in U87 cells transfected with si-DLEU1 or si-NC. The transwell invasion assay revealed that downregulation of DLEU1 significantly suppressed migration and invasion capacity of glioma cells (Figure 3A and B), suggesting that DLEU1 could affect glioma cell metastasis.
DLEU1 functions as a sponge for miR-421 in glioma
Accumulating evidence suggested that lncRNA could serve as a molecular sponge of miRNA to liberate the messenger RNA (mRNA) transcript targeted by miRNA.23 To investigate molecular mechanism that DLEU1 promoted cell glioma progression, we used StarBase 2.0 to select miRNA that binds to DLEU1. We found miR-421 with complementary sequences to the DLEU1 transcript (Figure 4A). In order to further investigate whether DLEU1 directly interacts with miR-421, luciferase reporter vectors containing wild type or mutated DLEU1 was constructed, and then luciferase reporter assay was also conducted. The results demonstrated that overexpression of miR-421 resulted in a significant reduction in luciferase activity of WT-DLEU1 in U87 cells (Figure 4B), but had no inhibitory effect on MT-DLEU1. To further verify the direct binding between miR-421 and DLEU1 in glioma cells, RIP assay was performed in U87 cells using the Ago2 antibody. The result showed that DLEU1 and miR-421 were specifically enriched in Ago2 pellets of U87 extracts compared to the IgG control group (Figure 4C). Moreover, we found that knockdown of DLEU1 led to a significant increase of miR-421 expression in U87 cells (Figure 4D), while overexpression of miR-421 decreased DLEU1 expression in U87 cells (Figure 4E). We also investigated the correlation with miR-421 and DLEU1 in glioma tissues and found that miR-421 expression was markedly downregulated in glioma tissues relative to adjacent normal brain tissues (Figure 4F). In addition, a negative correlation between DLEU1 and miR-421 expression was observed in glioma tissues (Figure 4G).
MEF2D has been reported to be a direct target of miR-421 in glioma.24 Growing evidence suggested that lncRNA could function as competing endogenous RNAs (ceRNAs) of miRNA, which would regulate their miRNA binding partners.25 Thus, to test association of DLEU1, miR-421 and MEF2D in glioma, we conducted the rescue assays by transfecting si-DLEU1 in combination with miR-421 inhibitor into U87 cells to examine MEF2D expression on mRNA and protein levels by quantitative real-time PCR and Western blot, respectively. The results revealed that downregulation of DLEUI reduced MEF2D expression on mRNA and protein levels in glioma cells, while the transfection with miR-421 inhibitor apparently abolished this trend (Figure 4H and I), suggesting that DLEU1 liberated MEF2D expression in glioma cells by competitively binding to miR-421. Furthermore, we found that MEF2D expression was increased in glioma tissues compared with adjacent normal brain tissues (Figure 4J), and its expression was positively correlated with DLEU1 expression (Figure 4K), and negatively correlated with miR-421 expression in glioma tissues (Figure 4L). Taken together, these findings suggested that DLEU1 could serve as a molecular sponge for miR-421 in glioma.
miR-421 inhibitor reverses the effects of DLEU1 knockdown in glioma cells
In order to test whether DLEU1 exerted its functional role in glioma cells through miR-421, we carried out the rescue experiments by transfecting si-DLEU1 plus miR-421 inhibitor into U87 cells. We found that downregulation of DLEU1 significantly impeded cell proliferation, induced cell apoptosis, and decreased migration and invasion in U87 cells, while the inhibition of miR-421 partially abrogated these effects mediated by DLEU1 knockdown in U87 cells (Figure 5A–D).
These results implied that DLEU1 exerted its functional roles in glioma cells via serving as a ceRNA for sponge miR-421.
Discussion
Many lncRNAs were reported to participate in the initiation and development of glomal and were considered as promising diagnostic biomarkers and therapeutic targets for glioma.8,9 For example, Meng et al reported that lncRNA Zinc Finger E-box-binding homeobox 1 antisense 1 (ZEB1-AS1) promoted glioma growth and metastasis by modulating the miR-200c/141-ZEB1 axis.26 Zhou et al demonstrated that lncRNAHOXD-AS1 promoted proliferation, migration and invasion and decreased cisplatin sensitivity of glioma cells by regulating miR-204.27 Zheng et al revealed that lncRNA AGAP2-AS1 drove glioma progression by sponging miR-15a/b-5p to regulate the expression of HDGF and activating Wnt/β-catenin signaling pathway.28 In the present study, we showed that the DLEU1 was significantly upregulated in glioma tissues and cell lines. Increased DLEU1 expression was correlated with glioma grade and KPS score. We also found that knockdown of DLEU1 inhibited tumorigenesis and development of glioma.
DLEU1 was reported to be frequently knocked down in multiple myeloma,29 chronic lymphocytic leukemia and other hematopoietic malignancies,19 and functioned as a tumor suppressor in these types of cancer. On the contrary, several studies have shown that DLEU1 functioned as an oncogenic lncRNA in multiple types of cancer.10–16 The contradictory role of DLEU1 in various tumors indicate that DLEU1 may exhibit different functional roles depending on various types of cancer. The role and underlying mechanism of DLEU1 in glioma remain obscure. Here, we found that DLEU1 expression was increased in glioma tissues compared with adjacent normal tissues. The higher DLEU1 is associated with glioma grade and KPS score. Our results revealed that that knockdown of DLEU1 suppressed the proliferation, promoted apoptosis, and decreased migration and invasion abilities of glioma cells. These results implied that DLEU1 played an oncogenic role in glioma progression.
Accumulating evidence suggests that lncRNA exhibits its biological function through competitive inhibition of miRNAs by serving as a molecular sponge.23,25 To explore the miRNA-related functions of DLEU1 in the pathogenesis of glioma, Starbase2.0 was used to select miRNAs that interact with DLEU1. miR-421, a known tumor suppressor, was used as a candidate miRNA for further investigation based on its biological function in glioma. Previously, a study demonstrated that miR-421 expression was downregulated and functioned as tumor suppressor in glioma.24 Here, through luciferase activity and RIP assays, we confirmed DLEU1 could bind to miR-421 in glioma cells. Quantitative real-time PCR analysis demonstrated that knockdown of DLEU1 increased miR-421 expression in U87 cells, while overexpression of miR-421 decreased DLEU1 expression in U87 cells. Furthermore, an inverse correlation between miR-421 expression and DLEU1 expression in glioma tissues was observed. Of note, inhibition of miR-421 partially abrogated the functional effect of DLEU1 knockdown on glioma cell proliferation, apoptosis, migration, and invasion. These data provided reliable evidence suggesting that DLEU1 knockdown exerts an inhibitory effect on glioma progression, at least in part, through targeting miR-421.
MEF2D, member of the myocyte enhancer factor 2 (MEF2) family of transcription factors, was reported to be upregulated and function as an oncogene in multiple types of cancers, such as lung cancer,30 gastric cancer,31 osteosarcoma32 and pancreatic cancer.33 Previously, a study reported that MEF2D expression was upregulated in malignant glioma tissues, and played a tumor-promoting role in glioma progression.34 Importantly, miR-421 inhibited glioma progression by targeting MEF2D.24 Here, we tested association DLEU1, miR-421 and MEF2D in glioma by rescue experiment. Our results revealed that downregulation of DLEUI reduced MEF2D expression on mRNA and protein levels in glioma cells, while the transfection with miR-421 inhibitor apparently abolished this trend. Furthermore, we found that MEF2D expression was positively correlated with DLEU1 expression and negatively correlated with miR-421 expression in glioma tissues. These results suggested that DLEU1 liberated MEF2D expression in glioma cells by competitively binding to miR-421, therapy resulting in promotion glioma progression.
In summary, the present study found that DLEU1 was up-regulated and played an oncogenic role in glioma by acting as a molecular sponge for miR-421 to regulate MEF2D expression. These findings suggested that DLEUI/miR-421/MEF2D molecular network played a vital role in glioma development and should be considered as a potential therapeutic target for glioma.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ordys BB, Launay S, Deighton RF, McCulloch J, Whittle IR. The role of mitochondria in glioma pathophysiology. Mol Neurobiol. 2010;42(1):64–75. doi:10.1007/s12035-010-8133-5
2. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
3. Groothuis DR. The blood-brain and blood-tumor barriers: a review of strategies for increasing drug delivery. Neuro-oncology. 2000;2(1):45–59. doi:10.1093/neuonc/2.1.45
4. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi:10.1016/j.cell.2014.03.008
5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
6. Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res. 2018;129:151–155. doi:10.1016/j.phrs.2017.11.009
7. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
8. Dang Y, Wei X, Xue L, Wen F, Gu J, Zheng H. Long non-coding RNA in glioma: target miRNA and signaling pathways. Clin Lab. 2018;64(6):887–894. doi:10.7754/Clin.Lab.2018.180107
9. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. lncRNAs as potential molecular biomarkers for the clinicopathology and prognosis of glioma: a systematic review and meta-analysis. Gene. 2018;668:77–86.
10. Li X, Li Z, Liu Z, Xiao J, Yu S, Song Y. Long non-coding RNA DLEU1 predicts poor prognosis of gastric cancer and contributes to cell proliferation by epigenetically suppressing KLF2. Cancer Gene Ther. 2018;25(3–4):58–67. doi:10.1038/s41417-017-0007-9
11. Wang LL, Sun KX, Wu DD, et al. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21(11):3055–3065. doi:10.1111/jcmm.13217
12. Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17(1):118. doi:10.1186/s12943-018-0873-2
13. Gao S, Cai Y, Zhang H, Hu F, Hou L, Xu Q. Long noncoding RNA DLEU1 aggravates pancreatic ductal adenocarcinoma carcinogenesis via the miR-381/CXCR4 axis. J Cell Physiol. 2019;234(5):6746–6757. doi:10.1002/jcp.27421
14. Shao W, Li Y, Chen F, Jia H, Jia J, Fu Y. Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression. Pharmazie. 2018;73(7):379–385. doi:10.1691/ph.2018.8352
15. Zhang S, Guan Y, Liu X, Ju M, Zhang Q. Long non-coding RNA DLEU1 exerts an oncogenic function in non-small cell lung cancer. Biomed Pharmacother. 2019;109:985–990. doi:10.1016/j.biopha.2018.10.175
16. Liu C, Tian X, Zhang J, Jiang L. Long non-coding RNA DLEU1 promotes proliferation and invasion by interacting with miR-381 and enhancing HOXA13 expression in cervical cancer. Front Genet. 2018;9:629. doi:10.3389/fgene.2018.00173
17. Nishiyama K, Maruyama R, Niinuma T, et al. Screening for long noncoding RNAs associated with oral squamous cell carcinoma reveals the potentially oncogenic actions of DLEU1. Cell Death Dis. 2018;9(8):826. doi:10.1038/s41419-018-0893-2
18. Li Y, Cui X, Li Y, Zhang T, Li S. Upregulated expression of miR-421 is associated with poor prognosis in non-small-cell lung cancer. Cancer Manag Res. 2018;10:2627–2633. doi:10.2147/CMAR.S167432
19. Lee S, Luo W, Shah T, et al. The effects of DLEU1 gene expression in Burkitt lymphoma (BL): potential mechanism of chemoimmunotherapy resistance in BL. Oncotarget. 2017;8(17):27839–27853. doi:10.18632/oncotarget.15711
20. Zhao J, Li B, Shu C, Ma Y, Gong Y. Downregulation of miR-30a is associated with proliferation and invasion via targeting MEF2D in cervical cancer. Oncol Lett. 2017;14(6):7437–7442. doi:10.3892/ol.2017.7114
21. Zhu Y, Zhao H, Feng L, Xu S. MicroRNA-217 inhibits cell proliferation and invasion by targeting Runx2 in human glioma. Am J Transl Res. 2016;8(3):1482–1491.
22. Araos J, Sleeman JP, Garvalov BK. The role of hypoxic signalling in metastasis: towards translating knowledge of basic biology into novel anti-tumour strategies. Clin Exp Metastasis. 2018;35(7):563–599.
23. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
24. Liu L, Cui S, Zhang R, Shi Y, Luo L. MiR-421 inhibits the malignant phenotype in glioma by directly targeting MEF2D. Am J Cancer Res. 2017;7(4):857–868.
25. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5). doi:10.3390/ijms19051310
26. Meng L, Ma P, Cai R, Guan Q, Wang M, Jin B. Long noncoding RNA ZEB1-AS1 promotes the tumorigenesis of glioma cancer cells by modulating the miR-200c/141-ZEB1 axis. Am J Transl Res. 2018;10(11):3395–3412.
27. Zhou H, Ma Y, Zhong D, Yang L. Knockdown of lncRNA HOXD-AS1 suppresses proliferation, migration and invasion and enhances cisplatin sensitivity of glioma cells by sponging miR-204. Biomed Pharmacother. 2019;112:108633. doi:10.1016/j.biopha.2019.108633
28. Zheng Y, Lu S, Xu Y, Zheng J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/beta-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530. doi:10.1016/j.ijbiomac.2019.01.121
29. Dowd AA, Homeida S, Elkarem HA. Detection of chromosome 13 (13q14) deletion among Sudanese patients with multiple myeloma using a molecular genetics fluorescent in situ hybridization technique (FISH). Malays J Pathol. 2015;37(2):95–100.
30. Zhang R, Zhang Y, Li H. miR-1244/myocyte enhancer factor 2D regulatory loop contributes to the growth of lung carcinoma. DNA Cell Biol. 2015;34(11):692–700. doi:10.1089/dna.2015.2915
31. Xu K, Zhao YC. MEF2D/Wnt/beta-catenin pathway regulates the proliferation of gastric cancer cells and is regulated by microRNA-19. Tumour Biol. 2016;37(7):9059–9069. doi:10.1007/s13277-015-4766-3
32. Du L, Chen T, Zhao K, Yang D. miR-30a suppresses osteosarcoma proliferation and metastasis by downregulating MEF2D expression. Onco Targets Ther. 2018;11:2195–2202. doi:10.2147/OTT.S102430
33. Song Z, Feng C, Lu Y, Gao Y, Lin Y, Dong C. Overexpression and biological function of MEF2D in human pancreatic cancer. Am J Transl Res. 2017;9(11):4836–4847.
34. Zhao Y, Li Y, Ma Y, et al. Myocyte enhancer factor 2D promotes tumorigenicity in malignant glioma cells. Tumour Biol. 2016;37(1):601–610. doi:10.1007/s13277-015-3791-6
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.