Back to Journals » Cancer Management and Research » Volume 11
Long noncoding RNA DANCR in various cancers: a meta-analysis and bioinformatics
Authors Wang L, Xie Y, Fang H, Zhang X , Pan H, Yan SX
Received 9 January 2019
Accepted for publication 10 June 2019
Published 15 July 2019 Volume 2019:11 Pages 6581—6592
DOI https://doi.org/10.2147/CMAR.S200922
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Lihong Wang,1 Yalin Xie,2 Hui Fang,1 Xia Zhang,1 Huiyun Pan,1 Senxiang Yan3
1Department of Geratology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, Zhejiang, People’s Republic of China; 2Department of Radiotherapy, Hangzhou Tumor Hospital, Hangzhou 310000, Zhejiang, People’s Republic of China; 3Department of Radiotherapy, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, Zhejiang, People’s Republic of China
Background: Differentiation antagonizing non-protein-coding RNA (DANCR) is a novel long noncoding RNA. Recent studies have shown that DANCR is aberrantly expressed in several types of cancer and is associated with poor outcomes. However, the clinical diagnostic significance of DANCR in tumors is not completely understood.
Methods: We searched the PubMed, Medline, Web of Science, EMBASE, Cochrane Library, and Ovid databases (up to December 30, 2018) for relevant literature. A total of 11 studies with 945 cancer patients were included in the present meta-analysis. We further validated the results using The Cancer Genome Atlas (TCGA) dataset.
Results: High expression of DANCR significantly predicted poor overall survival (low expression group vs high expression group; HR =0.56, 95% CI=[0.43, 0.72], =0.000); this was validated using TCGA. Moreover, DANCR expression was associated with advanced tumor node metastasis stage (I+II:III+IV; OR=0.22, 95% CI=[0.14, 0.35], P=0.001) and lymph node metastasis (no:yes; OR=0.21, 95% CI=[0.13, 0.35], P=0.001).
Conclusion: Our results suggest that elevated DANCR is related to poor clinical outcomes and could serve as a potential prognostic biomarker of cancer.
Keywords: cancer, overall survival, TCGA, long noncoding RNA, DANCR, meta-analysis
Introduction
Cancer is expected to rank as the leading cause of mortality and morbidity worldwide, with approximately 14 million new cases and 8 million cancer-related deaths every year, affecting populations in all countries and all regions.1,2 Despite the rapid development of science and technology, oncotherapy continues to face great challenges, including delayed diagnosis and poor prognosis, mainly due to the lack of specific biomarkers.3,4 Therefore, study of specific biomarkers in cancer, particularly the relationships between cancer progression and expression levels of biomarkers, may provide novel diagnostic methods and potential therapeutic targets for cancer.
Long noncoding RNAs (lncRNAs), a group of RNA molecules without protein-coding capacity and more than 200 nucleotides in length, can regulate gene expression at the transcriptional or post-transcriptional level.5,6 Increasing numbers of studies have implicated dysregulation of lncRNAs in cancers, including in proliferation, invasion, and metastasis, and shown that they can function as either tumor suppressors or oncogenes.7–10 Previous studies have demonstrated that 18% of lncRNAs are associated with tumors; much higher than the corresponding proportion (9%) of protein-coding genes.11 Moreover, some lncRNAs have been confirmed as specific prognostic biomarkers for cancers. Previous meta-analyses indicated a connection between expression levels of ZEB1-AS112 and PCAT-113 and prognosis of cancer. All these results suggest an important role for lncRNAs in carcinogenesis and cancer progression. They may thus represent promising novel prognostic biomarkers for tumors. LncRNA differentiation antagonizing non-protein-coding RNA (DANCR) is located on chromosome 4, with the closest adjacent annotated genes being USP46 and ERVMER34-1. DANCR was originally identified as a suppressor in the progression of epidermal cell differentiation,14 and it functions as an oncogenic driver in several types of cancer.15,16 Several studies have reported that DANCR is abnormally expressed in multiple tumors and associated with tumor proliferation, invasion, and metastasis. Further, there is increasing evidence that high expression levels of DANCR are related to clinicopathological features such as tumor size, lymph node metastasis (LNM), differentiation, tumor node metastasis (TNM) stage, and prognosis.16–26 These results have provided new insights into the role of DANCR in the development of cancers and indicated the potential applications of DANCR as a prognostic marker. However, these studies were limited in their ability to assess the implications of DANCR levels in cancer owing to inconsistent outcomes.20,21 To date, there has not been a meta-analysis of the published studies available. Therefore, in the pre sent study, we performed a correlation analysis based on meta-analysis and bioinformatics methods to investigate the relationship between DANCR and clinicopathological characteristics and prognosis.
Materials and methods
Literature search and study design
The study was performed according to standard guidelines for the meta-analysis of original studies.27,28 The PubMed, Medline, Web of Science, EMBASE, Cochrane Library, and Ovid databases were searched. The latest search time was December 30, 2018. The key words for searches were “DANCR” OR “Differentiation antagonizing non-protein coding RNA” OR “Long noncoding RNA DANCR” OR “lncRNA DANCR” and “Cancer” or “Tumor” or “Neoplasia” and “prognos*” or “outcome”.
Inclusion and exclusion criteria
Two researchers assessed all of the included studies and extracted the data independently. In this meta-analysis, the inclusion criteria were as follows: 1) the study reported original data and was published as a full peer-reviewed article; 2) the level of DANCR expression was measured in human tumor tissue by reverse-transcriptase polymerase chain reaction (RT-PCR) and patients were grouped according to different DANCR expression level; 3) the patients had a definitive diagnosis of cancer; 4) studies with sufficient original data for statistical analyses of survival information, namely, overall survival (OS) or clinicopathological features such as LNM, advanced TNM stage, or gender differences with lncRNA DANCR expression.
The exclusion criteria were as follows: 1) cell or animal experiments, letters, case reports, or reviews; 2) cases where the required data could not be extracted from the original article; 3) publications for which a later or more complete version was available for the same data subsets.
Data extraction and quality assessment
Data extraction from the original aritcles was conducted independently by two investigators for each included article. Any disagreements were resolved by discussion and consensus with a third investigator. The extracted data included the first author’s name, year of publication, country, method of DANCR testing, hazard ratio (HR) and the corresponding 95% confidence interval (CI) for lncRNA DANCR for OS (primary outcome) and clinicopathological parameters (gender, age, tumor size, differentiation, LNM, and TNM stage).A database was established after the selected data were arranged and verified. Quality assessment of the included studies was performed according to Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines28 (Table S1); the study scores ranged from 60% to 75%, and studies with more than 60% scores could be regarded as high quality. The detailed assessment was show in Table S2.
Statistical analysis
Engauge Digitizer 10.0 software was used to extract the survival data from Kaplan–Meier curves. STATA 12.0 software was used to analyze the extracted data. HR and corresponding 95% CI values were used to assess the association between DANCR and OS. Odds ratios (ORs) with 95% CIs were pooled to assess the correlation between DANCR expression and clinicopathological features. The chi-squared Q test and I2 statistics were used to evaluate the heterogeneity. A fixed‐effects model was chosen when there was no severe heterogeneity (I2<50% or P>0.1). Otherwise, a random-effects model was used. Begg’s test was used to evaluate potential publication bias, and sensitivity analyses were performed to examine the stability of the results.
Bioinformatics analysis
DANCR expression and clinical data of patients with cancer were retrieved from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). Htseq-counts data downloaded from TCGA were used to perform differential expression analysis between tumor samples and adjacent normal samples. The analysis was performed using the Edge R package;29 two-fold changes in expression levels and differences with false discovery rate <0.01 were considered significant. Survival analysis was performed using the median DANCR gene FPKM (fragments per kilobase of transcript per million fragments mapped) as a cutoff value; patients were classified into a DANCR high-expression group and a DANCR low-expression group.30 Kaplan–Meier survival curves and log-rank tests were used to assess the differences in survival time between the two group, using P<0.05 as a threshold. The analysis was performed using survival R packages.
Results
Study identification and characteristics
As shown in Figure 1, a total of 88 potential studies were collected from the database, of which 66 studies were removed as they were found to be non-DANCR-related, duplicated, or did not involve tests in tumor tissues when their titles and abstracts were reviewed; a total of 22 articles were identified for full review. Eleven studies were then excluded owing to lack of specific reporting on the association of DANCR with cancer or insufficient data in the original studies. Finally, the remaining 11 articles were included in the meta-analysis. The assessment for Risk of bias in individual studies is shown in Table S3.
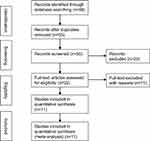 |
Figure 1 Flow diagram of the study search and selection process in the meta-analysis. |
As shown in Table 1, all reports were written in English and were published in China between 2015 and 2018. In these studies, a total of 945 patients were represented, with a mean sample size of 85.9 (range from 47 to 128). There were six types of cancer in this meta-analysis: urothelial carcinoma of the bladder, colorectal cancer, gastric cancer, glioma, nasopharyngeal carcinoma, non-small-cell lung cancer, and triple negative breast cancer. Patients were divided into two groups (high and low expression of DANCR). All the DANCR measurements were performed using RT-PCR. All measurements of pathological parameters were dependent on individual pathology.
 |
Table 1 Summary of the 11 included articles |
Association between DANCR and prognosis
The association between DANCR expression level and OS is shown in Figure 2. We performed a cumulative meta-analysis to assess the function of DANCR with respect to OS in cancer patients. Of the 11 included studies, seven studies with 667 patients reported on the relationship between OS and DANCR. A fixed-effects model was used because no obvious heterogeneity was found among those seven studies. The pooled HRs indicated that higher DANCR expression was related to worse survival (low DANCR expression group vs high expression group; pooled HR =0.56, 95% CI=[0.43, 0.72], P=0.000, fixed effect; Figure 2A). The stability of the pooled results was evaluated by sensitivity analysis (Figure 2B), which indicated that the results were reliable. Begg’s funnel plot was used to evaluate publication bias, showing that there was no publication bias for OS (P=0.133; Figure 2C).
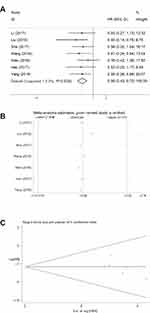 |
Figure 2 Forest plot of studies evaluating (A) the relationship between DANCR expression and overall survival (OS) rate, (B) sensitivity analysis for OS, and (C) Begg’s publication bias plots of OS. |
Association between DANCR and clinicopathologic characteristics
In this meta-analysis, four studies reporting on 325 patients with differentiation were included. As shown in Table 2, higher expression of DANCR was related to low differentiation (high + moderate:low, pooled OR=0.36, 95% CI=[0.15, 0.68], P=0.021, random effect). Further, the pooled ORs demonstrated that DANCR upregulation was related to LNM (no:yes, OR=0.21, 95% CI=[0.13, 0.35], P=0.001, fixed effect) and advanced TNM stage (I+II:III+IV, OR=0.22, 95% CI=[0.14, 0.35], P=0.001, fixed effect). However, no significant difference was observed between the two groups for tumor size (OR=0.72, 95% CI:=[0.36–1.42], P=0.354, random effect). The pooled results also demonstrated that the expression of DANCR was not related to gender (OR=1.29, 95% CI=[0.84, 1.96], P=0.754, fixed effect) or age (OR=1.27, 95% CI=[0.87, 1.86], P=0.219, fixed effect). Therefore, our results indicated that high DANCR expression significantly increased the risk of worse clinicopathological features. (Figure 3A–F). Begg’s funnel plot was used to evaluate publication bias (Figure 4A–F). There was no publication bias for gender (P=0.806), age (P=0.707), tumor size (P=0.806), differentiation (P=0.899), LNM (P=0.901), or TNM stage (P=0.221).
 |
Table 2 The association between DANCR expression and clinical features |
 |
Figure 3 Forest plot of studies evaluating the relationship between DANCR expression and (A) differentiation, (B) lymph node metastasis, (C) stage, (D) tumor size, (E) gender, and (F) age. |
 |
Figure 4 Begg’s publication bias plots evaluating the relationship between DANCR expression and (A) differentiation (B) lymph node metastasis (C) Stage (D) Tumor size (E) Gender (F) Age. |
Validation of the results in TCGA dataset
Bioinformatics analysis was performed to gain insight into the functional impact of the expression of DANCR on various cancers. We first evaluated the expression of DANCR in six different cancers using data from TCGA. As shown in Figure 5, DANCR was overexpressed in four of these cancers, including brain lower grade glioma, thymoma, diffuse large B-cell lymphoma, and cholangiocarcinoma (|Log2fold change (FC)| cutoff >1 and P<0.01). Kaplan–Meier curves with log-rank analysis were used to determine the relationships between the expression of DANCR and the OS rates of all patients with various cancers based on the TCGA dataset. Similar to the results of our meta-analysis, we found that higher DANCR expression (n=5021) was correlated with shorter survival time compared with patients with lower DANCR expression (n=5020) (log-rank P<0.05) (Figure 6A). The role of DANCR in different tumor types was then analyzed. Higher expression of DANCR was related to worse OS in adrenocortical carcinoma (ACC), liver hepatocellular carcinoma (LIHC), and sarcoma (SARC) (log-rank P<0.05) (Figure 6B–D). These results suggest that DANCR could function as an independent prognostic biomarker for tumors.
Discussion
Accumulating evidence has shown that DANCR is upregulated in various cancers. This meta-analysis was performed to clarify the association between DANCR and clinicopathologic features and prognosis in cancers. The results suggested that the risk of developing LNM, lower differentiation, and advanced TNM stage was higher in the high DANCR expression group compared with the low expression group. The pooled results also showed that high expression of DANCR was related to a worse OS; thus, DANCR could be an independent indicator for the prognosis of cancers. This meta-analysis is the first review to assess the relationship between DANCR and the prognosis of patients in various cancers. We further strengthened the results of our meta-analysis using bioinformatics methods. The Kaplan–Meier curves with log-rank analysis showed that, in ACC, LIHC and SKCM, the OS of patients with high DANCR expression was shorter than that of those with low DANCR expression. These results all illustrate that DANCR could function as a prognostic biomarker, with the potential for new cancer therapies that involve downregulation of DANCR expressions.
Previous studies also investigated the mechanism of DANCR in different cancers, with respect to cell proliferation, invasion, and metastasis. In glioma,18 DANCR markedly increased migration and proliferation of glioma cells by activating the Wnt/β-catenin signaling pathway. In hepatocellular carcinoma, Yuan et al15 found that DANCR increased the stemness features of tumor cells to promote tumorigenesis, invasion, and metastasis. In the respiratory system, DANCR contributes to the proliferation, migration, and invasion of lung adenocarcinoma cells by regulating miR-49631 and miR-758-3p.22 In bladder cancer, Zhan et al26 found that knockdown of DANCR inhibited epithelial-mesenchymal transition and malignant phenotypes of bladder cancer cells. DANCR promoted proliferation, migration, and invasion of cells in cervical cancer32 and osteosarcoma33 by acting as a competing endogenous RNA, regulating ROCK1 expression through binding to miR-335-5p.In colorectal cancer, Wang et al25 confirmed that overexpression of DANCR promoted proliferation and metastasis of tumor cells via miR-577 binding.
This meta-analysis had some limitations. First, all the studies included were from the People’s Republic of China, so our data may not represent global populations. Second, some HRs for OS were extracted from Kaplan–Meier curves, potentially leading to errors. Third, a limited number of tumor types and patients were included. More studies with larger sample sizes and examining various cancers should be included in future analyses.
Conclusions
Our meta-analysis suggested that high expression of DANCR was significantly associated with poor outcomes in various cancers and could serve as a potential prognostic biomarker. However, considering the limitations of the present analysis, more high-quality studies with large sample sizes are needed to confirm the prognostic value of DANCR in cancers.
Acknowledgments
This study was funded by the National Key R&D Program of China (No.2017YFC0907900/2017YFC0907904).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Stewart BW,Wild CP.The global and regional burden of cancer.In: World cancer report 2014. World Health Organization; 2015;16–54.
3. Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25(27):3866–3884. doi:10.1038/sj.onc.1209550
4. Desantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi:10.3322/caac.21235
5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
6. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
7. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No. 1):R17–R29. doi:10.1093/hmg/ddl046
8. Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi:10.1038/nature09144
9. Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21(11):1944–1954. doi:10.1101/gr.122358.111
10. Li T, Xie J, Shen C, et al. Amplification of long non-coding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. doi:10.1158/0008-5472.CAN-14-3569
11. Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124.11. doi:10.1016/j.canlet.2015.02.035
12. Liang C, Liu J, Ge H, Xu Y, Li G, Wu J. The clinicopathological and prognostic value of long non-coding RNA ZEB1-AS1 in solid tumors: a meta-analysis. Clin Chim Acta. 2018;484:91–98. doi:10.1016/j.cca.2018.05.011
13. Liang C, Qi Z, Ge H, et al. Long non-coding RNA PCAT-1 in human cancers: a meta-analysis. Clin Chim Acta. 2018;480:47–55. doi:10.1016/j.cca.2018.01.043
14. Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. doi:10.1101/gad.195123.112
15. Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi:10.1002/hep.27893
16. Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37:BSR20171070. doi:10.1042/BSR20171070
17. Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/β-catenin signaling. Biomed Pharmacother. 2018;102:602–607. doi:10.1016/j.biopha.2018.03.116
18. Liu Y, Zhang M, Liang L, et al. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480.
19. Hao YP, Qiu JH, Zhang DB, et al. Long non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancer. Tumor Biol. 2017;39(6):101042831769979. doi:10.1177/1010428317699798
20. Pan L, Liang W, Gu J, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9(2):1915–1930. doi:10.18632/oncotarget.23019
21. Sha S, Yuan D, Liu Y, et al. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):
22. Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi:10.1016/j.biopha.2018.03.053
23. Wen X, Liu X, Mao YP, et al. Long non-coding RNA DANCR stabilizes HIF-1α and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8(20):5676–5689. doi:10.7150/thno.28538
24. Yang JX, Sun Y, Gao L, et al. Long non-coding RNA DANCR facilitates glioma malignancy by sponging miR-33a-5p. Neoplasma. 2018;65:790–798. doi:10.4149/neo_2018_170724N498
25. Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50(5):57. doi:10.1038/s12276-018-0082-5
26. Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37(1). doi:10.1186/s13046-018-0921-1.
27. Fan YH, Ji CX, Xu B, et al. Long noncoding RNA activated by TGF-β in human cancers: a meta-analysis. Clin Chim Acta. 2017;468:10–16. doi:10.1016/j.cca.2017.02.001
28. Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 2012;9(5):e1001216. doi:10.1371/journal.pmed.1001216
29. Mccarthy DJ, Chen Y, Smyth GK, et al. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi:10.1093/nar/gks042
30. Dellinger TH, Smith DD, Ouyang C, et al.L1CAM is an independent predictor of poor survival in endometrial cancer - An analysis of The Cancer Genome Atlas (TCGA). Gynecol Oncol. 2016;141(2):336–340. doi:10.1016/j.ygyno.2016.02.003
31. Lu QC, Rui ZH, Guo ZL, et al. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med. 2018;22(3):1527–1537. doi:10.1111/jcmm.13420
32. Liang H, Zhang C, Guan H, et al. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. Cell Physiol. 2018. doi:10.1002/jcp.27484
33. Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46. doi:10.1016/j.canlet.2017.06.009
Supplementary materials
 |
Table S1 Reporting recommendations for tumor marker prognostic studies (REMARK) checklist |
 |
Table S2 Assessing the quality of included studies based on reporting recommendations for tumor marker prognostic studies (REMARK) guideline |
 |
Table S3 Risk of bias in individual studies |
References
1. Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/β-catenin signaling. Biomed Pharmacother. 2018;102:602–607. doi:10.1016/j.biopha.2018.03.116
2. Liu Y, Zhang M, Liang L, et al. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480.
3. Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37:BSR20171070. doi:10.1042/BSR20171070
4. Hao YP, Qiu JH, Zhang DB, et al. Long non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancer. Tumor Biol. 2017;39(6):101042831769979. doi:10.1177/1010428317699798
5. Pan L, Liang W, Gu J, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9(2):1915–1930. doi:10.18632/oncotarget.23019
6. Sha S, Yuan D, Liu Y, et al. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):
7. Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi:10.1016/j.biopha.2018.03.053
8. Wen X, Liu X, Mao YP, et al. Long non-coding RNA DANCR stabilizes HIF-1α and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8(20):5676–5689. doi:10.7150/thno.28538
9. Yang JX, Sun Y, Gao L, et al. Long non-coding RNA DANCR facilitates glioma malignancy by sponging miR-33a-5p. Neoplasma. 2018;65:790–798. doi:10.4149/neo_2018_170724N498
10. Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50(5):57. doi:10.1038/s12276-018-0082-5
11. Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37(1). doi:10.1186/s13046-018-0921-1.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


