Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers
Authors Qin Y, Sun W, Wang Z, Dong W , He L, Zhang T, Zhang H
Received 11 June 2020
Accepted for publication 17 July 2020
Published 5 August 2020 Volume 2020:13 Pages 7699—7717
DOI https://doi.org/10.2147/OTT.S267140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Yuan Qin ,* Wei Sun ,* Zhihong Wang, Wenwu Dong, Liang He, Ting Zhang, Hao Zhang
Department of Thyroid Surgery, The First Hospital of China Medical University, Shenyang 110001, Liaoning Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hao Zhang
Department of Thyroid Surgery, The First Hospital of China Medical University, 155 Nanjing Bei Street, Shenyang, Liaoning 110001, People’s Republic of China
Email [email protected]
Abstract: Long non-coding RNAs (lncRNAs) are emerging regulators of a diverse range of biological processes through various mechanisms. Genome-wide association studies of tumor samples have identified several lncRNAs, which act as either oncogenes or tumor suppressors in various types of cancers. Small nucleolar RNAs (snoRNAs) are predominantly found in the nucleolus and function as guide RNAs for the processing of transcription. As the host genes of snoRNAs, lncRNA small nucleolar RNA host genes (SNHGs) have been shown to be abnormally expressed in multiple cancers and can participate in cell proliferation, tumor progression, metastasis, and chemoresistance. Here, we review the biological functions and emerging mechanisms of SNHGs involved in the development and progression of endocrine-related cancers including thyroid cancer, breast cancer, pancreatic cancer, ovarian cancer and prostate cancer.
Keywords: endocrine, cancers, lncRNA, SNHG
Introduction
Long non-coding RNAs (lncRNAs, >200 nucleotides in length) are emerging regulators of gene transcription.1 The human genome estimated to encode >28,000 lncRNAs,2 but only 15,778 lncRNAs are annotated in the current GENECODE version 27.3 Therefore, more lncRNAs are yet to be discovered. Moreover, the known lncRNAs have not been studied in depth.
Accumulating evidence suggests lncRNAs play key roles in the development and progression of several cancers, acting as either oncogenes or tumor suppressors.4 LncRNAs can regulate transcription, translation, protein modification, and the formation of RNA-protein or protein-protein complexes, depending on the cellular location.5 For example, lncRNAs primarily located in the nucleus are involved in transcriptional regulation and mRNA processing, while cytoplasmic lncRNAs play roles in modulating mRNA translation by competing with proteins or in miRNA-mediated mRNA decoy.5,6
Small nucleolar RNAs (snoRNAs, 60–300 nucleotides in length) are more well-characterized than lncRNAs and are predominantly found in the nucleolus.7 Most snoRNAs function as guide RNAs for the post-transcriptional modification of ribosomal RNAs and some spliceosomal RNAs, with some involved in the nucleolytic processing of the original rRNA transcript.8 As shown in Figure 1, the majority of snoRNAs are encoded (hosted) in the introns of protein-coding and non-protein-coding genes, termed small nucleolar RNA host genes (SNHGs).9–11 Primary RNA transcripts of host genes (including all exons and introns with their snoRNAs) are cut into different exons and introns. Exons are then re-spliced and function in the cytoplasm, while the introns are further processed into snoRNAs and play roles in the nucleolus.
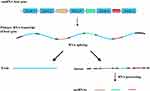 |
Figure 1 The synthetic pathway of snoRNAs. |
Currently, there are 22 members of SNHG family (SNHG1 to SNHG22) that have been shown to regulate proliferation, apoptosis, invasion, and migration in multiple cancers, including endocrine-related cancers (as summarized in Tables 1 and 2). These 22 SNHGs have diverse activities and mechanisms of action. For example, SNHG1 has been shown to promote colorectal cancer cell growth by modulating histone methylation of gene promoters of the Kruppel Like Factor 2 (KLF2, a member of the KLF family, also exerts tumor-suppressive roles) and the cyclin-dependent kinase 4 inhibitor B (CDKN2B, a tumor suppressor).12 SNHG1 can also act as a sponge for miR-154-5p to upregulate expression of G1/S-specific cyclin-D2 (CCND2, which is involved in cell cycle progression).12 Meanwhile, SNHG13 serves as a competing endogenous RNA (ceRNA) of miR-34a-5p, leading to the derepression of Jagged 1 (JAG1) expression, which eventually triggers resistance to docetaxel in prostate cancer.13
 |
Table 1 Characteristics of SNHG Members |
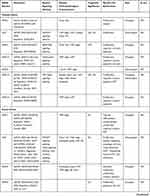 | 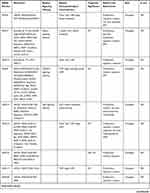 | 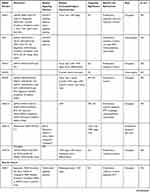 | 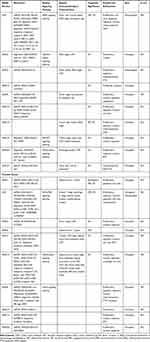 |
Table 2 SNHG Members in Endocrine-Related Cancers |
This review aims to provide an overview on the current understanding of the regulation and function of SNHGs in endocrine-related cancers that arise from the endocrine glands or neuroendocrine tissues, including thyroid cancer, breast cancer, pancreatic cancer, ovarian cancer, and prostate cancer.14
Thyroid Cancer
Thyroid cancer is the most common malignancy of the endocrine system with enormous heterogeneity in terms of morphological features and prognosis.15 Although the majority of cases of thyroid cancer tend to be biologically indolent and have an excellent prognosis, some are associated with more aggressive clinical behavior.16
SNHG1 may act as an oncogene in thyroid cancer by competing with miR-199a-5p and upregulating the expression of its target gene, the transcription factor (TF) SP1. In turn, SP1 targets the promoter region of SNHG1 and promote its transcription, forming a positive feedback loop to promote cancer cell proliferation and invasion.17 Conversely, low expression of SNHG2, also known as growth arrest specific transcript 5 (GAS5), is associated with poor prognosis of patients with thyroid cancer.18 Mechanistically, GAS5 acts as a sponge for miR-222-3p, thereby modulating the expression of the phosphatase and tensin homolog (PTEN), leading to PTEN/protein kinase B (AKT) pathway activation and the suppression of thyroid cancer cell proliferation.19
SNHG7 is also markedly upregulated in thyroid cancer samples, with high SNHG7 expression associated with shorter survival times.20 Indeed, SNHG7 knockdown leads to a suppression of thyroid cancer cell proliferation and migration, and induction of apoptosis via downregulating the acyl-CoA synthetase long chain family member 1 (ACSL1) and the brain-derived neurotrophic factor (BDNF).21,22 In addition, bioinformatics analysis showed SNHG7 was associated with the processes of “protein translation”, “viral life cycle”, “RNA processing”, “mRNA splicing”, “histone ubiquitination”, “endoplasmic reticulum-to-Golgi vesicle-mediated transport”, “sister chromatid cohesion”, “DNA damage checkpoint regulation”, “translation”, and “the spliceosome”, suggesting further research directions for this lncRNA.20
SNHG12 is also upregulated (by 3.8-fold) in papillary thyroid carcinoma (PTC) tissues compared to normal adjacent tissue samples.23 High SNHG12 was associated with poorer progression in PTC in terms of tumor node metastasis (TNM) staging and lymph node metastasis (LNM).24 SNHG12 likely acts as a sponge for miR-16-5p, thereby inducing PTC cell proliferation, migration, and invasion, as well as inhibiting apoptosis.25 SNHG12 also promotes the proliferation and migration of PTC cells via the Wnt/β-catenin signaling pathway.23 Meanwhile, SNHG13, also known as differentiation antagonizing non-protein coding RNA (DANCR), acts as a tumor suppressor in PTC: downregulation of DANCR is associated with more aggressive clinical features of PTC.26 DANCR is also a potential biomarker for PTC diagnosis, showing a sensitivity of 85.29% and a specificity of 66.18%.26
The role of SNHG15 in thyroid cancer remains controversial. SNHG15 is upregulated in human PTC tissues and cell lines compared to controls, and was associated with gender, larger tumor size, LNM, advanced TNM stage, and poorer overall survival (OS).27 Meanwhile, SNHG15 downregulation attenuated cell proliferation, migration, and epithelial–mesenchymal transition (EMT) in PTC cells, as well as inducing apoptosis.27 Mechanistically, SNHG15 acts as a sponge for miR-200a-3p, thereby upregulating the Yes-associated protein 1 (YAP1) signaling pathway.27 Alternatively, another study showed SNHG15 was downregulated in thyroid cancer tissues and cell lines and suppressed tumor progression, indicating SNHG15 may act as a tumor suppressor.28 Moreover, inhibition of SNHG15 by miR-510-5p promoted cell proliferation, migration, and invasion in thyroid cancer.29 These diverse functions of SNHG15 found in different studies may reflect the different subtypes of thyroid cancer; however, further research is required.
Finally, SNHG16, which functions as an endogenous sponge for miR-497, was upregulated in both PTC tissues and cell lines and shown to induce proliferation, migration, and invasion of thyroid cancer cells, while inhibiting apoptosis.30 High expression of SNHG16 was also positively associated with advanced TNM stage and LNM.30
In summary, SNHG1, GAS5, SNHG7, SNHG12, DANCR, SNHG15, and SNHG16 all appear to play essential roles in thyroid cancer; although the function of SNHG15 requires further confirmation.
Breast Cancer
Breast cancer is the most commonly diagnosed cancer worldwide and the leading cause of cancer-related death for women.31 Although advances in early detection and cancer therapeutics have led to a decrease in mortality rates, breast cancer remains a significant public health concern. Some classes of breast cancer, such as triple-negative breast cancer (characterized by a lack of expression of the progesterone receptor, estrogen receptor, and Her-2), have a poor prognosis.32 Many lncRNAs have been implicated in breast cancer development in recent years, which may eventually lead to better outcomes for these patients.33
The downregulation of SNHG1 can suppress the proliferation and invasion of breast cancer cells by regulating miR-382.34 In addition, SNHG1 may inhibit the differentiation of regulatory T cells, promote miR-448 expression, and reduce indoleamine 2,3 dioxygenase (IDO) levels in breast cancer.35 Therefore, SNHG1 may be a useful target in breast cancer treatment.
GAS5 was first reported to be a tumor suppressor in breast cancer in 2009.36 Since then, studies have shown low GAS5 expression is closely related to a more aggressive tumor phenotype, enhanced proliferation, and attenuated apoptosis in breast cancer cells.37–39 GAS5 can bind to miR-196a-5p, thereby partially alleviating its tumor-promoting effects, including invasion and downstream forkhead box O1 (FOXO1)/phosphatidylinositol 3-kinase (PI3K)/AKT signal pathway activation.37 Notch-1 also promotes breast cancer cell proliferation by downregulating GAS5.40 GAS5 can also act as a sponge for miR-23a to promote autophagy via the GAS5-miR-23a-ATG3 axis in breast cancer.38 Moreover, in drug-resistant breast cancer cells, GAS5 overexpression increases chemosensitivity (eg to trastuzumab, imatinib, paclitaxel, cisplatin, among others), especially in triple-negative breast cancer cells.39,41-46 Another study showed miR-221/222 suppresses GAS5 expression and enhances tumor growth in a mouse model of breast cancer xenografts.47 Moreover, lower plasma GAS5 levels were found in patients with a high Ki67 proliferation index before surgery and in those with LNM after surgery.48 Finally, bioinformatics analysis showed GAS5 plays a role in “proliferation” and the “cell cycle”, although the molecular mechanisms related to these regulatory pathways are unclear.49
There is evidence that lncRNA secreted in exosomes from cancer cells can regulate gene expression and signaling pathways in other niche cells. For example, breast cancer-derived cancer-associated fibroblasts can secrete increased amounts of SNHG3 than healthy breast tissue cells, which in turn promotes the growth of breast cancer cells by regulating miR-330-5p/Pyruvate Kinase M1/M2 (PKM).50 SNHG3 can also act as a sponge for miR-384/hepatoma-derived growth factor (HDGF) to drive breast cancer cell proliferation, migration, and invasion.51
SNHG5 is an oncogene and acts as a sponge for miR-154-5p, reducing its ability to repress proliferating cell nuclear antigen (PCNA), thus promoting breast cancer proliferation, cell cycle progression, and inhibiting apoptosis.52 SNHG6 was also found to be highly expressed in breast cancer tissues and cell lines, and is associated with poorer clinicopathologic features.53 Indeed, SNHG6 knockdown inhibits breast cancer cell proliferation, migration, invasion, and G1 cell cycle arrest by acting as a sponge for miR-26a-5p, which regulates expression of the vasodilator-stimulated phosphoprotein (VASP)54 and mitogen-activated protein kinase 6 (MAPK6).55
The expression of SNHG7 is also upregulated in breast cancer tissues and cells compared to healthy tissues, with high SNHG7 expression strongly related to tumor stage, distant metastasis, LNM, and OS.56–58 Knocking down SNHG7 inhibited breast cancer cell proliferation, invasion, and EMT.56–58 Further mechanistic studies revealed SNHG7 could act as a sponge to repress miR-34a,57 miR-186,58 and miR-381,56 thereby activating the Notch-1 pathway and glycolysis in breast cancer. Additionally, c-Myc (a TF) can bind to the SNHG7 promoter and positively regulate its expression in breast cancer.59
Increased expression of SNHG12 has been observed in triple-negative breast cancer.60 SNHG12 upregulation positively correlated with advanced tumor stage and size, and negatively correlated with OS.60 SNHG12 is a direct transcriptional target of c-Myc, and the c-Myc-induced upregulation of SNHG12 enhances the proliferation of breast cancer cells and inhibits apoptosis.60 SNHG12 may also promote the migration of breast cancer cells by regulating the expression of matrix metalloproteinase 13 (MMP13).60
High DANCR levels can lead to shorter OS in triple-negative breast cancer, by acting as a sponge for miR-216a-5p and thereby promoting the proliferation and invasion of tumor cells.61 DANCR can mediate protein assembly and modification in triple-negative breast cancer. For example, DANCR can bind to the phosphorylation site of retinoid X receptor alpha (RXRA) and suppresses its interaction with the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) promoter.62 This leads to the activation of the P13K/AKT pathway, which in turn, promotes the proliferation and growth of triple-negative breast cancer cells.62 DANCR may also participate in the enhancer of zeste homolog 2 (EHZ2)-mediated epigenetic repression of the suppressor of cytokine signaling 3 (SOCS3) in breast cancer cells.63 Sha et al64 proposed DANCR knockdown was associated with increased binding of EZH2 to the promoters of CD44 and ABCG2 (two triple-negative breast cancer stem cell markers), and the concomitant reduction of expression of these genes decreased cancer cell proliferation and invasion. Furthermore, nanoparticle-mediated RNAi of DANCR was shown to be an effective therapy for triple-negative breast cancer.65
Upregulation of SNHG14 in breast cancer tissues may also promote cancer cell proliferation and invasion.66 In particular, SNHG14 upregulates polyadenylate-binding protein 1 (PABPC1) expression by modulating H3K27 acetylation (H3K27ac) in the promoter of PABPC1 gene, resulting in the activation of the nuclear factor E2-related factor 2 (NRF2) signaling pathway, which is involved in cell defense and survival against chemotherapy drugs.66 Besides histone methylation, acetylation is another important form of histone modification.
Indeed, exosomal SNHG14 was upregulated in trastuzumab-resistant human epidermal growth factor receptor 2 (HER2) breast cancer cells compared with parental breast cancer cells, and SNHG14 knockdown re-sensitized breast cancer cells to trastuzumab treatment.67 These results indicate SNHG14 may be a promising therapeutic target for patients with HER2+ breast cancer. In addition, SNHG14 may enhance breast cancer cell proliferation and invasion by acting as a sponge for miR-193a-3p.68
SNHG15 has also been shown to be highly expressed in breast cancer tissues and cell lines and is positively associated with larger tumor size, LNM, advanced TMN stage, and worse survival.69,70 SNHG15 primarily acts as a sponge for miR-411-5p69 and miR-211-3p,70 leading to the proliferation, migration, and invasion of breast cancer cells. Additionally, SNHG15 knockdown enhances the cisplatin sensitivity of breast cancer cells by acting as a sponge for miR-381.71 Moreover, bioinformatics analysis showed SNHG16 might be associated with the prognosis of breast cancer.72,73 In particular, SNHG16 may interact with miR-30a to regulate the expression of ribonucleoside-diphosphate reductase subunit M2 (RRM2)74 and competitively bind miR-98 and the E2F Transcription Factor 5 (E2F5)75 to promote the proliferation and invasion of breast cancer cells. Finally, SNHG1776 and SNHG2077 may also drive breast cancer progression by sponging miR-124-3p and miR-495, respectively.
In general, multiple SNHGs, including SNHG1, GAS5, SNHG3, SNHG5, SNHG6, SNHG7, SNHG12, DANCR, SNHG14, SNHG15, SNHG16 and SNHG20, play a role in breast cancer. Targeting SNHGs, especially the treatment of drug-resistant breast cancer, is the future research direction.
Pancreatic Cancer
Pancreatic cancer is one of the most devastating human tumors, with high invasiveness, early metastasis, lack of specific symptoms, and high mortality. According to the most recent statistical data, the 5-year survival of pancreatic cancer is 9%, which is the lowest among all types of cancers and continues to increase (by 0.3% per year) in men.78 The high fatality rate in pancreatic cancer is attributed to late diagnosis and resistance to current therapies. Recent studies demonstrate lncRNAs are critical in the pathogenesis of pancreatic cancer and are therefore potential biomarkers or drug targets.79
SNHG1 acts as an oncogene in pancreatic cancer and accelerates cancer cell growth.80 In addition, SNHG1 overexpression can promote cyclin D1-mediated pancreatic cancer proliferation by regulating the cell cycle.81 Meanwhile, SNHG1 downregulation inhibits the proliferation, migration, and invasion of pancreatic cancer cells by suppressing the Notch-1 signaling pathway.80 Similarly, SNHG1 downregulation inhibits the PI3K/AKT signaling pathway in pancreatic ductal adenocarcinoma (PDAC).82
By acting as a sponge for miR-32-5p, GAS5 can promote the expression of PTEN and stop the activation of the PI3K/AKT signaling pathway, thereby inhibiting pancreatic cancer cell proliferation and survival.83 GAS5 also inhibits the expression of the oncogene cyclin-dependent kinase 6 (CDK6), although the underlying mechanisms have not been determined.84 Studies also show GAS5 reduces the chemoresistance of pancreatic cancer cells by downregulating miR-181c-5p and miR-221.85,86
SNHG7 is highly expressed in pancreatic cancer tissues and positively correlates with reduced OS. Meanwhile, SNHG7 knockdown suppresses cell proliferation, migration, and invasion of pancreatic cancer cells by modulating the miR-342-3p/inhibitor of DNA binding 4 (ID4) axis.87 Zhang et al88 showed low expression of SNHG9 in pancreatic cancer tissues and serums, while those with high SNHG9 expression had significantly higher survival rates. This data indicates SNHG9 may be a novel prognostic marker for pancreatic cancer.
High DANCR expression correlates with vascular invasion, advanced T grade, LNM, and advanced TNM stage, and is an independent risk factor for poor OS and progression-free survival (PFS) in PDAC.89,90 Mechanistically, DANCR acts as an miRNA sponge, affecting the miRNA-33a-5p/Anexelekto (AXL) axis,89 the miRNA-33b/MMP16 axis,91 the miR-135a/NLRP3 axis,92 and the miR-214-5p/E2F2 axis90 to promote cell proliferation, migration, invasion, and metastasis in pancreatic cancer.
The SNHG14 oncogene also potentiates pancreatic cancer cell proliferation through modulation of annexin A2 (ANXA2) expression by acting as a ceRNA for miR-613.93 It also acts as a sponge for miR-10, thereby enhancing autophagy, which underlies the chemoresistance of PDAC cells to gemcitabine.94,95 Finally, SNHG15 and SNHG16 are upregulated in pancreatic cancer samples and are associated with progression in pancreatic cancer patients.96,97 SNHG15 may help repress P15 and KLF2 expression,96 while SNHG16 promotes cell proliferation, migration, and invasion of pancreatic cancer by sponging miR-200a-3p98 and miR-218-5p.97 SNHG16 may also promote pancreatic cancer lipogenesis by directly regulating the miR-195/SREBP2 axis.99
In short, many SNHGs have a significant predictive effect on the survival of pancreatic cancer patients, and can be used as a clinical prognostic marker in pancreatic cancer.
Ovarian Carcinoma
Ovarian cancer is the most lethal gynecological cancer in women globally.100 Despite recent improvements in cytoreductive surgery and chemotherapy, the 5-year survival rate of ovarian cancer is still approximately 40–50% owing to its late diagnosis and the development of chemoresistance.78 Therefore, understanding the molecular mechanisms of ovarian carcinogenesis may help improve diagnosis, therapy, and prevention.
Expression of SNHG1 is increased in human epithelial ovarian cancer tissues and cell lines compared to normal healthy tissues, and promotes the proliferation and invasion of ovarian carcinoma cells through the regulation of EMT and the Wnt/β-catenin pathway.101,102 Meanwhile, GAS5 acts as a tumor suppressor and is expressed in low levels epithelial ovarian cancer samples.103,104 Indeed, GAS5 expression correlates with prognosis in epithelial ovarian cancer, including International Federation of Gynecology and Obstetrics (FIGO) stage, histological type, OS, and disease-free survival (DFS).103,104 In terms of its mechanism of action, GAS5 may block CCAAT/enhancer-binding protein beta (CEBPB)-mediated transcription of the growth/differentiation factor 15 (GD15), leading to decreased viability and increased apoptosis of ovarian cancer cells.105 GAS5 may also suppress the proliferation of ovarian cancer cells by sponging miR-21106 and miR-196a-5p,107 which regulate sprouty homolog 2 (SPRY2) and homeobox A5 (HOXA5) expression, respectively. GAS5 is also implicated in inflammasome formation and pyroptosis, but the underlying mechanism is unclear.108 Finally, GAS5 has been linked to chemoresistance; in particular, GAS5 overexpression control the expression of poly(ADP-ribose) polymerase 1 (PARP1) by recruiting the transcription factor E2F4 to its promoter, which subsequently affects the mitogen-activated protein kinase (MAPK) pathway activity, further enhance the cisplatin sensitivity of ovarian cancer cells.109
Upregulation of SNHG3 expression is associated with poor prognosis in ovarian cancer (including FIGO stage and LNM) and promotes proliferation and invasion by activating the GSK3β/-catenin signaling pathway.110 Bioinformatics analysis has shown SNHG3 is related to energy metabolism in the “glycolysis”, “Kreb’s cycle”, and “oxidative phosphorylation” pathways, and to “drug resistance”.111 Similarly, SNHG5 has been implicated in chemoresistance: paclitaxel-resistant ovarian cancer tissues and cell lines have lower levels of SNHG5, while SNHG5 overexpression can enhance paclitaxel sensitivity (likely by sponging miR-23a).112
SNHG12 is also upregulated in ovarian cancer tissues and enhances the proliferative and migratory capacity of cells via sponging miR-129 and upregulating expression of SOX4 (a TF).113 In addition, DANCR levels are higher in ovarian cancer patients with worse tumor stage and accompanied by metastatic loci.114 DANCR binds directly to miR-145 and regulates vascular endothelial growth factor (VEGF) expression.115 Indeed, knockdown of DANCR impairs ovarian tumor growth by inhibiting tumor angiogenesis.115 In addition, DANCR may enhance the proliferation, migration, and invasion capacities of ovarian cancer cells by upregulating expression of the insulin-like growth factor 2 (IGF2)116 and downregulating UPF1 RNA Helicase And ATPase (UPF1) expression.114
Like SNHG12, SNHG14 is highly expressed in ovarian cancer tissues and associated with poorer OS.117,118 SNHG14 may promote ovarian cancer cell progression by sponging miR-125a-5p117 and miR-219a-5p,118 or directly regulating the expression of DiGeorge syndrome chromosomal region 8 (DGCR8).119 SNHG15 and SNHG16 may also serve as oncogenes in epithelial ovarian cancer. SNHG16 has been shown to promote the proliferation, invasion, and migration of cancer cells via activation of the PI3K/AKT signaling pathway,120 while the role of SNHG15 is unclear.121 SNHG20 is also upregulated in ovarian cancer and is associated with shorter OS.122 SNHG20 knockdown suppresses Wnt/β-catenin signaling activity and EMT-associated gene expression, thereby inhibiting ovarian cancer cell proliferation, migration, and invasion.123 Finally, the SNHG22 oncogene may regulate the miR-2467/Gal-1 axis to promote cisplatin- and paclitaxel-resistance of ovarian cancer cells.124
In a word, compared with other SNHGs, GAS5 regulates the progression of ovarian cancer through various mechanisms, indicating its key role in the development of ovarian cancer.
Prostate Cancer
Prostate cancer is the most common malignancy in males and accounts for 10% of cancer-related deaths.78 Androgen deprivation therapy (ADT) is the standard treatment for patients with biochemical recurrence after primary treatment, or with locally-advanced or metastatic disease. However, the majority of cancers will eventually acquire ADT resistance and progress to castration-resistant prostate cancer (CRPC).125 Aberrantly expressed lncRNAs can be indicative of certain stages of prostate cancer progression, and may predict early progression or efficiently sustain tumor‐related signaling pathways. Thus, lncRNAs may be applicable for the diagnosis of prostate cancer, as well as being potential criteria in the choice of therapy and new therapeutic targets of CRPC.126
SNHG1 upregulation in prostate cancer correlates with the Gleason score, T stage, and a short biochemical recurrence-free survival time.127 SNHG1 may promote prostate cancer cell proliferation by regulating the miR-199a-3p/CDK7 axis128 and the miR-377-3p/AKT2 axis.129 Conversely, GAS5 levels are reduced in prostate cancer tissues and cell lines.130–132 Low GAS5 levels are associated with prostate-specific antigen level, Gleason score, and pathological stage.130–132
Most studies indicate that GAS5 inhibits the proliferation, migration, and invasion of prostate cancer cells, and promotes apoptosis.130–132 In terms of its mechanism of action, GAS5 may act as a sponge for miR-103, which in turn, inactivates the AKT/mTOR signaling pathway, thus inhibiting prostate cancer cell proliferation.131 In addition, two single nucleotide polymorphisms (SNPs) located in the chromosomal segment of GAS5 (rs55829688 and rs145204276) can increase GAS5 expression,133–135 and are associated with improved survival in prostate cancer.133 Patients with prostate cancer and the GAS5 rs145204276 polymorphism are associated with a low risk of pathologic N stage and seminal vesicle invasion.135 Furthermore, patients with prostate cancer aged >65 years who carry the GAS5 rs145204276 polymorphism show decreased risk of clinical T stage, pathologic N stage, and lymphovascular invasion.135 Differential expression of GAS5 due to these SNPs likely affects the miR-21/programmed cell death 4 (PDCD4)/PTEN axis,133 as well as the miR-1284/AKT133 and miR-1284/high mobility group box 1 (HMGB1)134 pathways. In addition, overexpression of miR-145 can upregulate GAS5 expression, although GAS5 overexpression (or silencing) has no effect on miR-145 levels.132
Enhancing GAS5 expression may be particularly useful in androgen-sensitive prostate cancers.136 Indeed, mTOR inhibitors enhance GAS5 transcript levels in androgen-sensitive prostate cancer cell lines but have no effect on androgen-independent cell lines (which exhibit low endogenous levels of GAS5).136 As further evidence of its tumor-suppressing role, GAS5 is implicated in improving the radiosensitivity of prostate cancer cells. In particular, GAS5 can enhance the α-Solanine-induced radiosensitivity of prostate cancer cells by negatively regulating miR-18a.137
Despite available evidence showing that GAS5 acts as a tumor suppressor, some studies report GAS5 may exist as an oncogene in prostate cancer. For example, Zhang and Chen et al.138,139 found GAS5 expression was higher in prostate cancer tissues than normal healthy tissues in both public databases and human tissue samples. In addition, functional analysis showed GAS5 knockdown inhibited the proliferation and cell cycle progression of prostate cancer cells, while promoting apoptosis.138 A bioinformatics analysis also showed high expression of GAS5 correlated with poorer DFS in prostate cancer, and other studies show GAS5 may be involved in regulating translational elongation, protein biosynthesis, transcription, protein translation, and proliferation.138–140
SP1-mediated upregulation of SNHG4 can facilitate prostate cancer progression via the miR-377/zic family member 5 (ZIC5) axis.141 Similarly, SNHG6 overexpression was associated with shorter DFS in the Cancer Genome Atlas (TCGA) and Taylor datasets, with bioinformatics analysis revealing SNHG6 is associated with “translation”, “nuclear-transcribed mRNA catabolic processes”, “ribosomal RNA processing”, and “mRNA splicing”.142 SNHG7 is also significantly upregulated in prostate cancer tissue and cell lines,143,144 and correlates with the Gleason score, bone metastasis, pelvic LNM, TNM stage, and OS.145 In terms of its mechanism of action, SNHG7 knockdown was found to inhibit proliferation and promote CCND1-induced cell cycle arrest at the G0/G1 phase.144 SNHG7 can also promote EMT via regulating miR-324-3p and WNT2B, an important protein in the Wnt signaling pathway.143 Therefore, targeting the SNHG7/miR-324-p/WNT2B axis may represent a novel therapeutic strategy for prostate cancer treatment.
As SNHG12 acts as an oncogene, it may be a useful predictor of poor prognosis in prostate cancer. Indeed, a study showed SNHG12 acts as a sponge for miR-195 and can activate the Wnt/β-catenin signaling pathway.146 SNHG12 can also promote cell viability and inhibit apoptosis and autophagy of prostate cancer cells via regulating the expression of the G1/S-specific cyclin-E1 (CCNE1) by sponging miR-195.147 Bioinformatic analysis revealed higher expression of SNHG12 was enriched in the “P53 signaling pathway”, “cell cycle”, “regulation of cell migration”, “cellular metabolic process”, “gene expression”, and “Notch signaling pathway”, and that SNHG12 may target miR-133b.148
The oncogene DANCR has also been shown to promote the invasion and migration of prostate cancer cells in vitro and the metastasis of tumor xenografts in nude mice.149 Mechanistically, DANCR works synergistically with EZH2 to downregulate the expression of the tissue inhibitor of metalloproteinases (TIMP) 2/3.149 Furthermore, downregulation of DANCR can increase the paclitaxel sensitivity of prostate cancer cells by negatively regulating the expression of miR-135a.150 In addition, stimulation of the DANCR/miR-34a-5p axis enhanced docetaxel-resistance in prostate cancer via targeting JAG1, which in turn activates the Notch signaling pathway.13 Finally, SNHG14,151 SNHG15,152 and SNHG20153 may all act as oncogenes in prostate cancer via targeting miR-613, miR-338-3p, and miR-6516-5p to promote cell proliferation, migration, and invasion.
In conclusion, SNHGs plays an important role in the process and embody diversified treatment strategies in prostate cancer, especially in CRPC.
Conclusion
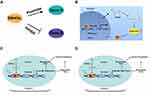
This review highlights that the abnormal expression of SNHGs is significantly related to poor prognosis (eg TNM stage, LNM, OS, DFS) and function (eg proliferation, invasion, migration, apoptosis, autophagy, and chemoresistance) in multiple endocrine-related cancers. Some SNHGs played similar roles in different tumors. For example, SNHG1, SNHG3, SNHG4, SNHG6, SNHG7, SNHG12, SNHG14, SNHG16, SNHG17, SNHG20 and SNHG22 promotes tumor growth as oncogenes, while GAS5 and SNHG9 played the role of tumor suppressor genes. In addition, SNHG5, DANCR, SNHG15 played a dual role, which have attracted more scholars’ attention. SNHGs could regulate the tumor process via various mechanisms, including direct regulation (promotion or inhibition) (Figure 2A), binding and being activated by TFs, acting as a ceRNA, activating different signaling pathways (Figure 2B), and regulating promoter methylation (Figure 2C) or acetylation of downstream target genes (Figure 2D). Both methylation and acetylation were histone modifications and their mechanisms were similar. The difference between them was that they bound and modified different histones, and then promoted or inhibited the expression of downstream genes. However, the SNHGs described in this review are only just the tip of the iceberg, and further mechanistic will be required as more SNHG family members are uncovered.
Abbreviations
ACSL1, acyl-CoA synthetase long chain family member 1; ADT, androgen deprivation therapy; AKT, protein kinase B; ANXA2, annexin A2; AXL, Anexelekto; BDNF, brain-derived neurotrophic factor; CCND1/2, cyclin-D1, cyclin-D2; CCNE1, cyclin-E1; CDK6/7, cyclin-dependent kinase 6, cyclin-dependent kinase 7; CDKN2B, cyclin-dependent kinase 4 inhibitor B; CEBPB, CCAAT/enhancer-binding protein beta; CeRNA, competing endogenous RNA; CRPC, castration-resistant prostate cancer; DANCR, Differentiation antagonizing non-protein coding RNA; DFS, disease-free survival; DGCR8, DiGeorge syndrome chromosomal region 8; E2F5, E2F Transcription Factor 5; EHZ2, enhancer of zeste homolog 2; EMT, epithelial–mesenchymal transition; FIGO, International Federation of Gynecology and Obstetrics; FOXO1, forkhead box O1; GAS5, growth arrest specific transcript 5; GD15, growth/differentiation factor 15; HDGF, hepatoma-derived growth factor; HER2, human epidermal growth factor receptor 2; HMGB1, high mobility group box 1; HOXA5, homeobox A5; IDO, indoleamine 2,3 dioxygenase; IGF2, insulin-like growth factor 2; JAG1, Jagged 1; KLF2, Kruppel Like Factor 2; LncRNA, long non-coding RNA; LNM, lymph node metastasis; MAPK6, mitogen-activated protein kinase 6; MMP13, matrix metalloproteinase 13; NRF2, nuclear factor E2-related factor 2; OS, overall survival; PABPC1, polyadenylate-binding protein 1; PARP1, poly(ADP-ribose) polymerase 1; PCNA, proliferating cell nuclear antigen; PDAC, pancreatic ductal adenocarcinoma; PDCD4, programmed cell death 4; PFS, progression-free survival; PI3K, phosphatidylinositol 3-kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PKM, pyruvate Kinase M1/M2; PTC, papillary thyroid carcinoma; PTEN, phosphatase and tensin homolog; RRM2, ribonucleoside-diphosphate reductase subunit M2; RXRA, retinoid X receptor alpha; SNHG, small nucleolar RNA host genes; SnoRNA, small nucleolar RNA; SNP, single nucleotide polymorphisms; SOCS3, suppressor of cytokine signaling 3; SPRY2, sprouty homolog 2; TCGA, the Cancer Genome Atlas; TF, transcription Factor; TIMP, tissue inhibitor of metalloproteinases; TNM, tumor node metastasis; UPF1, UPF1 RNA Helicase And ATPase; VASP, vasodilator-stimulated phosphoprotein; VEGF, Vascular endothelial growth factor; YAP1, Yes-associated protein 1; ZIC5, zic family member 5.
Data Sharing Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Qin Y, Sun W, Zhang H, et al. LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer. Endocrine. 2018;59(3):555–564. doi:10.1007/s12020-017-1520-1
2. Tragante V, Moore JH, Asselbergs FW. The ENCODE project and perspectives on pathways. Genet Epidemiol. 2014;38(4):275–280. doi:10.1002/gepi.21802
3. Klinge CM. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer. 2018;25(4):R259–R282. doi:10.1530/ERC-17-0548
4. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
5. Tang Y, Cheung BB, Atmadibrata B, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017;391:12–19. doi:10.1016/j.canlet.2017.01.010
6. Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14(2):73–80. doi:10.1016/j.gpb.2016.03.005
7. Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34(Database issue):D158–62. doi:10.1093/nar/gkj002
8. Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9(3):337–342. doi:10.1016/S0955-0674(97)80005-1
9. Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379(6564):464–466. doi:10.1038/379464a0
10. Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5ʹ-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi:10.1128/MCB.18.12.6897
11. Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer. Nat Rev Cancer. 2012;12(2):84–88. doi:10.1038/nrc3195
12. Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141. doi:10.1186/s12943-018-0894-x
13. Ma Y, Fan B, Ren Z, Liu B, Wang Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. Onco Targets Ther. 2019;12:5485–5497. doi:10.2147/OTT.S197009
14. Anik A, Abaci A. Endocrine cancer syndromes: an update. Minerva Pediatr. 2014;66(6):533–547.
15. Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–580. doi:10.1038/nrendo.2011.142
16. LiVolsi VA. Papillary thyroid carcinoma: an update. Mod Pathol. 2011;24(Suppl 2):S1–9. doi:10.1038/modpathol.2010.129
17. Ding W, Zhao S, Shi Y, Chen S. Positive feedback loop SP1/SNHG1/miR-199a-5p promotes the malignant properties of thyroid cancer. Biochem Biophys Res Commun. 2020;522(3):724–730. doi:10.1016/j.bbrc.2019.11.075
18. Guo LJ, Zhang S, Gao B, et al. Low expression of long non-coding RNA GAS5 is associated with poor prognosis of patients with thyroid cancer. Exp Mol Pathol. 2017;102(3):500–504. doi:10.1016/j.yexmp.2017.05.008
19. Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. 2018;9(3):3519–3530. doi:10.18632/oncotarget.23336
20. Chen L, Zhu J, Zhang LJ. Long non-coding RNA small nucleolar RNA host gene 7 is upregulated and promotes cell proliferation in thyroid cancer. Oncol Lett. 2019;18(5):4726–4734. doi:10.3892/ol.2019.10782
21. Wang YH, Huo BL, Li C, Ma G, Cao W. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci. 2019;23(11):4815–4821. doi:10.26355/eurrev_201906_18067
22. Guo L, Lu J, Gao J, Li M, Wang H, Zhan X. The function of SNHG7/miR-449a/ACSL1 axis in thyroid cancer. J Cell Biochem. 2020. doi:10.1002/jcb.29569
23. Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/β-catenin signaling pathway. Cancer Biomark. 2018;22(2):217–226. doi:10.3233/CBM-170777
24. Liu J, Tang X, Lv J, et al. LncRNAs SNHG12 and LINC00152 were associated with progression of patients with papillary thyroid carcinoma. Future Oncol. 2019;15(36):4167–4179. doi:10.2217/fon-2019-0016
25. Feng X, Dong X, Wu D, Zhao H, Xu C, Li H. Long noncoding RNA small nucleolar RNA host gene 12 promotes papillary thyroid carcinoma cell growth and invasion by targeting miR-16-5p. Histol Histopathol. 2020;35(2):217–224. doi:10.14670/HH-18-155
26. Zhang K, Lv J, Peng X, et al. Down-regulation of DANCR acts as a potential biomarker for papillary thyroid cancer diagnosis. Biosci Rep. 2019;39(4).
27. Wu DM, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. doi:10.1038/s41419-018-0975-1
28. Liu Y, Li J, Li F, Li M, Shao Y, Wu L. SNHG15 functions as a tumor suppressor in thyroid cancer. J Cell Biochem. 2019;120(4):6120–6126. doi:10.1002/jcb.27899
29. Liu Y, Li J, Li M, Li F, Shao Y, Wu L. microRNA-510-5p promotes thyroid cancer cell proliferation, migration, and invasion through suppressing SNHG15. J Cell Biochem. 2019.
30. Wen Q, Zhao L, Wang T, et al. LncRNA SNHG16 drives proliferation and invasion of papillary thyroid cancer through modulation of miR-497. Onco Targets Ther. 2019;12:699–708. doi:10.2147/OTT.S186923
31. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
32. Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223(2):307–317. doi:10.1002/path.2808
33. Lo PK, Wolfson B, Zhou X, Duru N, Gernapudi R, Zhou Q. Noncoding RNAs in breast cancer. Brief Funct Genomics. 2016;15(3):200–221. doi:10.1093/bfgp/elv055
34. Zheng S, Li M, Miao K, Xu H. SNHG1 contributes to proliferation and invasion by regulating miR-382 in breast cancer. Cancer Manag Res. 2019;11:5589–5598. doi:10.2147/CMAR.S198624
35. Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. 2018;118(Pt A):24–30. doi:10.1016/j.ijbiomac.2018.06.033
36. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi:10.1038/onc.2008.373
37. Li S, Zhou J, Wang Z, Wang P, Gao X, Wang Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother. 2018;104:451–457. doi:10.1016/j.biopha.2018.05.056
38. Gu J, Wang Y, Wang X, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48(1):194–207. doi:10.1159/000491718
39. Li J, Li L, Yuan H, Huang XW, Xiang T, Dai S. Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle. 2019;18(16):1965–1975. doi:10.1080/15384101.2019.1635870
40. Pei J, Wang B. Notch-1 promotes breast cancer cells proliferation by regulating LncRNA GAS5. Int J Clin Exp Med. 2015;8(8):14464–14471.
41. Pickard MR, Williams GT. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res Treat. 2014;145(2):359–370. doi:10.1007/s10549-014-2974-y
42. Esmatabadi M, Motamedrad M, Sadeghizadeh M. Down-regulation of lncRNA, GAS5 decreases chemotherapeutic effect of dendrosomal curcumin (DNC) in breast cancer cells. Phytomedicine. 2018;42:56–65. doi:10.1016/j.phymed.2018.03.022
43. Li W, Zhai L, Wang H, et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7(19):27778–27786. doi:10.18632/oncotarget.8413
44. Gu J, Wang Y, Wang X, et al. Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett. 2018;434:1–10. doi:10.1016/j.canlet.2018.06.039
45. Zheng S, Li M, Miao K, Xu H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J Cell Biochem. 2020;121(3):2225–2235. doi:10.1002/jcb.29445
46. Chen Z, Pan T, Jiang D, et al. The lncRNA-GAS5/miR-221-3p/DKK2 axis modulates ABCB1-mediated adriamycin resistance of breast cancer via the Wnt/β-catenin signaling pathway. Mol Ther Nucleic Acids. 2020;19:1434–1448. doi:10.1016/j.omtn.2020.01.030
47. Zong Y, Zhang Y, Sun X, Xu T, Cheng X, Qin Y. miR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA GAS5 in breast cancer. Biosci Rep. 2019;39(1). doi:10.1042/BSR20181859
48. Han L, Ma P, Liu SM, Zhou X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol. 2016;37(5):6847–6854. doi:10.1007/s13277-015-4568-7
49. Tokgun PE, Tokgun O, Kurt S, Tomatir AG, Akca H. MYC-driven regulation of long non-coding RNA profiles in breast cancer cells. Gene. 2019;714:143955. doi:10.1016/j.gene.2019.143955
50. Li Y, Zhao Z, Liu W, Li X. SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Appl Biochem Biotechnol. 2020.
51. Ma Q, Qi X, Lin X, Li L, Chen L, Hu W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33(1):232–242. doi:10.1007/s13577-019-00287-9
52. Chi JR, Yu ZH, Liu BW, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA Axis. Mol Ther Nucleic Acids. 2019;17:138–149. doi:10.1016/j.omtn.2019.05.013
53. Jafari-Oliayi A, Asadi MH. SNHG6 is upregulated in primary breast cancers and promotes cell cycle progression in breast cancer-derived cell lines. Cell Oncol (Dordr). 2019;42(2):211–221. doi:10.1007/s13402-019-00422-6
54. Li K, Ma YB, Tian YH, et al. Silencing lncRNA SNHG6 suppresses proliferation and invasion of breast cancer cells through miR-26a/VASP axis. Pathol Res Pract. 2019;215(10):152575. doi:10.1016/j.prp.2019.152575
55. Lv P, Qiu X, Gu Y, Yang X, Xu X, Yang Y. Long non-coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR-26a-5p/MAPK6 in breast cancer. Biomed Pharmacother. 2019;110:294–301. doi:10.1016/j.biopha.2018.11.016
56. Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci. 2019;23(15):6588–6595. doi:10.26355/eurrev_201908_18545
57. Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the notch-1 pathway. Eur J Pharmacol. 2019;856:172407. doi:10.1016/j.ejphar.2019.172407
58. Luo X, Song Y, Tang L, Sun DH, Ji DG. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. 2018;22(22):7788–7797. doi:10.26355/eurrev_201811_16403
59. Zhang L, Fu Y, Guo H. c-Myc-induced long non-coding RNA small nucleolar RNA host gene 7 regulates glycolysis in breast cancer. J Breast Cancer. 2019;22(4):533–547. doi:10.4048/jbc.2019.22.e54
60. Wang O, Yang F, Liu Y, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9(2):533–545.
61. Tao W, Wang C, Zhu B, Zhang G, Pang D. LncRNA DANCR contributes to tumor progression via targetting miR-216a-5p in breast cancer: lncRNA DANCR contributes to tumor progression. Biosci Rep. 2019;39(4). doi:10.1042/BSR20181618
62. Tang J, Zhong G, Zhang H, et al. LncRNA DANCR upregulates PI3K/AKT signaling through activating serine phosphorylation of RXRA. Cell Death Dis. 2018;9(12):1167. doi:10.1038/s41419-018-1220-7
63. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14(2):309–328. doi:10.1002/1878-0261.12622
64. Sha S, Yuan D, Liu Y, Han B, Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol Open. 2017;6(9):1310–1316. doi:10.1242/bio.023135
65. Vaidya AM, Sun Z, Ayat N, et al. Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triple-negative breast cancer therapy. Bioconjug Chem. 2019;30(3):907–919. doi:10.1021/acs.bioconjchem.9b00028
66. Dong H, Wang W, Mo S, et al. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J Cell Mol Med. 2018;22(10):4935–4947. doi:10.1111/jcmm.13758
67. Dong H, Wang W, Chen R, et al. Exosome-mediated transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol. 2018;53(3):1013–1026. doi:10.3892/ijo.2018.4467
68. Xie SD, Qin C, Jin LD, et al. Long noncoding RNA SNHG14 promotes breast cancer cell proliferation and invasion via sponging miR-193a-3p. Eur Rev Med Pharmacol Sci. 2019;23(6):2461–2468. doi:10.26355/eurrev_201903_17393
69. Liu LB, Jiang ZJ, Jiang XL, Wang S. Up-regulation of SNHG15 facilitates cell proliferation, migration, invasion and suppresses cell apoptosis in breast cancer by regulating miR-411-5p/VASP axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1899–1912. doi:10.26355/eurrev_202002_20368
70. Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun. 2018;495(2):1594–1600. doi:10.1016/j.bbrc.2017.12.013
71. Mi H, Wang X, Wang F, et al. SNHG15 contributes to cisplatin resistance in breast cancer through sponging miR-381. Onco Targets Ther. 2020;13:657–666. doi:10.2147/OTT.S223321
72. Li J, Gao C, Liu C, et al. Four lncRNAs associated with breast cancer prognosis identified by coexpression network analysis. J Cell Physiol. 2019;234(8):14019–14030. doi:10.1002/jcp.28089
73. Zhong G, Lou W, Yao M, Du C, Wei H, Fu P. Identification of novel mRNA-miRNA-lncRNA competing endogenous RNA network associated with prognosis of breast cancer. Epigenomics. 2019;11(13):1501–1518. doi:10.2217/epi-2019-0209
74. Du SM. The SNHG16/miR-30a axis promotes breast cancer cell proliferation and invasion by regulating RRM2. Neoplasma. 2020;67(03):567–575. doi:10.4149/neo_2020_190625N550
75. Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. doi:10.1016/j.bbrc.2017.02.094
76. Du Y, Wei N, Hong J, Pan W. Long non-coding RNASNHG17 promotes the progression of breast cancer by sponging miR-124-3p. Cancer Cell Int. 2020;20(1):40. doi:10.1186/s12935-020-1129-y
77. Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu SZ, Zhang YL. Lnc RNA SNHG20 participated in proliferation, invasion, and migration of breast cancer cells via miR-495. J Cell Biochem. 2018;119(10):7971–7981. doi:10.1002/jcb.26588
78. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
79. Saluja A, Maitra A. Pancreatitis and pancreatic cancer. Gastroenterology. 2019;156(7):1937–1940. doi:10.1053/j.gastro.2019.03.050
80. Cui L, Dong Y, Wang X, et al. Downregulation of long noncoding RNA SNHG1 inhibits cell proliferation, metastasis, and invasion by suppressing the notch-1 signaling pathway in pancreatic cancer. J Cell Biochem. 2019;120(4):6106–6112. doi:10.1002/jcb.27897
81. Li D, Zhang X, Yang Y, et al. Long non-coding RNA SNHG1 promotes cyclin D1-mediated proliferation in pancreatic cancer by acting as a ceRNA of miR-195. Int J Clin Exp Pathol. 2019;12(3):730–739.
82. Zhang Y, Zhang R, Luo G, Ai K. Long noncoding RNA SNHG1 promotes cell proliferation through PI3K/AKT signaling pathway in pancreatic ductal adenocarcinoma. J Cancer. 2018;9(15):2713–2722. doi:10.7150/jca.26207
83. Gao ZQ, Wang JF, Chen DH, et al. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017;7(1):66. doi:10.1186/s13578-017-0192-0
84. Lu X, Fang Y, Wang Z, et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354(3):891–896. doi:10.1007/s00441-013-1711-x
85. Gao Z-Q, Wang J-F, Chen D-H, et al. Long non-coding RNA GAS5 antagonizes the chemoresistance of pancreatic cancer cells through down-regulation of miR-181c-5p. Biomed Pharmacother. 2018;97:809–817. doi:10.1016/j.biopha.2017.10.157
86. Liu B, Wu S, Ma J, et al. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi:10.1016/j.omtn.2018.09.026
87. Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9(1):28. doi:10.1186/s13578-019-0290-2
88. Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10(8):2648–2658.
89. Chen L, Liu J, Tang T, et al. lncRNA differentiation antagonizing nonprotein coding RNA overexpression accelerates progression and indicates poor prognosis in pancreatic ductal adenocarcinoma. Onco Targets Ther. 2018;11:7955–7965. doi:10.2147/OTT.S167065
90. Yao Z, Chen Q, Ni Z, et al. Long non-coding RNA differentiation antagonizing nonprotein coding RNA (DANCR) promotes proliferation and invasion of pancreatic cancer by sponging miR-214-5p to regulate E2F2 expression. Med Sci Monit. 2019;25:4544–4552. doi:10.12659/MSM.916960
91. Luo Y, Wang Q, Teng L, et al. LncRNA DANCR promotes proliferation and metastasis in pancreatic cancer by regulating miRNA-33b. FEBS Open Bio. 2020;10(1):18–27. doi:10.1002/2211-5463.12732
92. Tang Y, Cao G, Zhao G, Wang C, Qin Q. LncRNA differentiation antagonizing non-protein coding RNA promotes proliferation and invasion through regulating miR-135a/NLRP37 axis in pancreatic cancer. Invest New Drugs. 2019.
93. Deng PC, Chen WB, Cai HH, et al. LncRNA SNHG14 potentiates pancreatic cancer progression via modulation of annexin A2 expression by acting as a competing endogenous RNA for miR-613. J Cell Mol Med. 2019;23(11):7222–7232. doi:10.1111/jcmm.14467
94. Zhang X, Zhao P, Wang C, Xin B. SNHG14 enhances gemcitabine resistance by sponging miR-101 to stimulate cell autophagy in pancreatic cancer. Biochem Biophys Res Commun. 2019;510(4):508–514. doi:10.1016/j.bbrc.2019.01.109
95. Xie F, Huang Q, Wang C, et al. Downregulation of long noncoding RNA SNHG14 suppresses cell proliferation and invasion by regulating EZH2 in pancreatic ductal adenocarcinoma (PDAC). Cancer Biomark. 2020;27(3):357–364. doi:10.3233/CBM-190908
96. Ma Z, Huang H, Wang J, et al. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget. 2017;8(48):84153–84167. doi:10.18632/oncotarget.20359
97. Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862. doi:10.1016/j.biopha.2019.108862
98. Guo JQ, Yang ZJ, Wang S, Wu ZZ, Yin LL, Wang DC. LncRNA SNHG16 functions as an oncogene by sponging miR-200a-3p in pancreatic cancer. Eur Rev Med Pharmacol Sci. 2020;24(4):1718–1724. doi:10.26355/eurrev_202002_20347
99. Yu Y, Dong JT, He B, et al. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15(33):3831–3844. doi:10.2217/fon-2019-0321
100. Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23(Suppl 10):x111–7. doi:10.1093/annonc/mds300
101. Ge J, Wu XM, Yang XT, Gao JM, Wang F, Ye KF. Role of long non-coding RNA SNHG1 in occurrence and progression of ovarian carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(2):329–335. doi:10.26355/eurrev_201801_14176
102. Wang S, Jiang J, Wang Z, Xie Y, Wu X. Long non-coding RNA SNHG1 is an unfavorable prognostic factor and promotes cell proliferation and migration by Wnt/β-catenin pathway in epithelial ovarian cancer. Int J Clin Exp Pathol. 2017;10(9):9284–9292.
103. Gao J, Liu M, Zou Y, et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep. 2015;34(6):3212–3221. doi:10.3892/or.2015.4318
104. Li J, Huang H, Li Y, Li L, Hou W, You Z. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol Rep. 2016;36(6):3241–3250. doi:10.3892/or.2016.5200
105. Guo LL, Wang SF. Downregulated long noncoding RNA GAS5 fails to function as decoy of CEBPB, resulting in increased GDF15 expression and rapid ovarian cancer cell proliferation. Cancer Biother Radiopharm. 2019;34(8):537–546. doi:10.1089/cbr.2019.2889
106. Ma N, Li S, Zhang Q, Wang H, Qin H, Wang S. Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Exp Ther Med. 2018;16(1):73–82. doi:10.3892/etm.2018.6188
107. Zhao H, Yu H, Zheng J, et al. Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol Oncol. 2018;151(2):345–355. doi:10.1016/j.ygyno.2018.08.032
108. Li J, Yang C, Li Y, Chen A, Li L, You Z. LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. 2018;38(2).
109. Long X, Song K, Hu H, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38(1):345. doi:10.1186/s13046-019-1329-2
110. Hong L, Chen W, Wu D, Wang Y. Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomark. 2018;22(3):367–374. doi:10.3233/CBM-170710
111. Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150(2):343–354. doi:10.1016/j.ygyno.2018.06.013
112. Lin H, Shen L, Lin Q, et al. SNHG5 enhances Paclitaxel sensitivity of ovarian cancer cells through sponging miR-23a. Biomed Pharmacother. 2020;123:109711. doi:10.1016/j.biopha.2019.109711
113. Sun D, Fan XH. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing miRNA-129 to upregulate SOX4. Eur Rev Med Pharmacol Sci. 2019;23(6):2345–2352. doi:10.26355/eurrev_201903_17378
114. Pei CL, Fei KL, Yuan XY, Gong XJ. LncRNA DANCR aggravates the progression of ovarian cancer by downregulating UPF1. Eur Rev Med Pharmacol Sci. 2019;23(24):10657–10663. doi:10.26355/eurrev_201912_19763
115. Lin X, Yang F, Qi X, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol Carcinog. 2019;58(12):2286–2296. doi:10.1002/mc.23117
116. Gao YQ, Cheng HY, Liu KF. Long non-coding RNA DANCR upregulates IGF2 expression and promotes ovarian cancer progression. Eur Rev Med Pharmacol Sci. 2019;23(9):3621–3626. doi:10.26355/eurrev_201905_17785
117. Zhao YL, Huang YM. LncSNHG14 promotes ovarian cancer by targeting microRNA-125a-5p. Eur Rev Med Pharmacol Sci. 2019;23(8):3235–3242. doi:10.26355/eurrev_201904_17683
118. Li L, Zhang R, Li SJ. Long noncoding RNA SNHG14 promotes ovarian cancer cell proliferation and metastasis via sponging miR-219a-5p. Eur Rev Med Pharmacol Sci. 2019;23(10):4136–4142. doi:10.26355/eurrev_201905_17915
119. Zhao JL, Wang CL, Liu YL, Zhang GY. Long noncoding RNA SNHG14 enhances migration and invasion of ovarian cancer by upregulating DGCR8. Eur Rev Med Pharmacol Sci. 2019;23(23):10226–10233. doi:10.26355/eurrev_201912_19659
120. Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(3):616–622.
121. Qu C, Dai C, Guo Y, Qin R, Liu J. Long noncoding RNA SNHG15 serves as an oncogene and predicts poor prognosis in epithelial ovarian cancer. Onco Targets Ther. 2019;12:101–111. doi:10.2147/OTT.S182657
122. Wang D, Dai J, Hou S, Qian Y. LncRNA SNHG20 predicts a poor prognosis and promotes cell progression in epithelial ovarian cancer. Biosci Rep. 2019;39(4).
123. He S, Zhao Y, Wang X, et al. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/β-catenin signaling. Biosci Rep. 2018;38(1). doi:10.1042/BSR20170681.
124. Zhang PF, Wu J, Luo JH, et al. SNHG22 overexpression indicates poor prognosis and induces chemotherapy resistance via the miR-2467/Gal-1 signaling pathway in epithelial ovarian carcinoma. Aging (Albany NY). 2019;11(19):8204–8216. doi:10.18632/aging.102313
125. Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017;108(11):2107–2114.
126. Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract. 2011;65(11):1180–1192. doi:10.1111/j.1742-1241.2011.02799.x
127. Wan X, Huang W, Yang S, et al. Identification of androgen-responsive lncRNAs as diagnostic and prognostic markers for prostate cancer. Oncotarget. 2016;7(37):60503–60518. doi:10.18632/oncotarget.11391
128. Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–152. doi:10.1016/j.bbrc.2017.03.169
129. Xie M, Zhang Z, Cui Y. Long noncoding RNA SNHG1 contributes to the promotion of prostate cancer cells through regulating miR-377-3p/AKT2 axis. Cancer Biother Radiopharm. 2020;35(2):109–119. doi:10.1089/cbr.2019.3177
130. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832(10):1613–1623. doi:10.1016/j.bbadis.2013.05.005
131. Xue D, Zhou C, Lu H, Xu R, Xu X, He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016;37(12):16187–16197. doi:10.1007/s13277-016-5429-8
132. Xie X, Dai J, Huang X, Fang C, He W. MicroRNA-145 inhibits proliferation and induces apoptosis in human prostate carcinoma by upregulating long non-coding RNA GAS5. Oncol Lett. 2019;18(2):1043–1048. doi:10.3892/ol.2019.10419
133. Zhu L, Zhu Q, Wen H, Huang X, Zheng G. Mutations in GAS5 affect the transformation from benign prostate proliferation to aggressive prostate cancer by affecting the transcription efficiency of GAS5. J Cell Physiol. 2019;234(6):8928–8940.
134. Deng ZH, Yu GS, Pan B, et al. Rs145204276 and rs4759314 affect the prognosis of prostate cancer by modulating the GAS5/miR-1284/HMGB1 and HOTAIR/miR-22/HMGB1 signalling pathways. Artif Cells Nanomed Biotechnol. 2020;48(1):435–442. doi:10.1080/21691401.2019.1709859
135. Lin CY, Wang SS, Yang CK, et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. Int J Med Sci. 2019;16(11):1424–1429. doi:10.7150/ijms.38080
136. Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75(7):693–705. doi:10.1002/pros.22952
137. Yang J, Hao T, Sun J, Wei P, Zhang H. Long noncoding RNA GAS5 modulates α-Solanine-induced radiosensitivity by negatively regulating miR-18a in human prostate cancer cells. Biomed Pharmacother. 2019;112:108656. doi:10.1016/j.biopha.2019.108656
138. Zhang Y, Su X, Kong Z, et al. An androgen reduced transcript of LncRNA GAS5 promoted prostate cancer proliferation. PLoS One. 2017;12(8):e0182305. doi:10.1371/journal.pone.0182305
139. Chen X, Yang C, Xie S, Cheung E. Long non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in translation targeted by miR-940 in prostate cancer. Oncotarget. 2018;9(1):1048–1062. doi:10.18632/oncotarget.23254
140. Romanuik TL, Wang G, Morozova O, Delaney A, Marra MA, Sadar MD. LNCaP atlas: gene expression associated with in vivo progression to castration-recurrent prostate cancer. BMC Med Genomics. 2010;3:43. doi:10.1186/1755-8794-3-43
141. Wang ZY, Duan Y, Wang P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol. 2020;235(4):3916–3927. doi:10.1002/jcp.29285
142. Yan Y, Chen Z, Xiao Y, Wang X, Qian K. Long non-coding RNA SNHG6 is upregulated in prostate cancer and predicts poor prognosis. Mol Biol Rep. 2019;46(3):2771–2778. doi:10.1007/s11033-019-04723-9
143. Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol Res Pract. 2019;215(10):152537. doi:10.1016/j.prp.2019.152537
144. Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332. doi:10.1016/j.biopha.2018.03.011
145. Xia Q, Li J, Yang Z, Zhang D, Tian J, Gu B. Long non-coding RNA small nucleolar RNA host gene 7 expression level in prostate cancer tissues predicts the prognosis of patients with prostate cancer. Medicine (Baltimore). 2020;99(7):e18993. doi:10.1097/MD.0000000000018993
146. Song J, Wu X, Ma R, Miao L, Xiong L, Zhao W. Long noncoding RNA SNHG12 promotes cell proliferation and activates Wnt/β-catenin signaling in prostate cancer through sponging microRNA-195. J Cell Biochem. 2019;120(8):13066–13075. doi:10.1002/jcb.28578
147. Wang X, He C, Yang Z, Li S, Qiao L, Fang L. Dysregulation of long non-coding RNA SNHG12 alters the viability, apoptosis, and autophagy of prostate cancer cells by regulating miR-195/CCNE1 axis. Int J Clin Exp Pathol. 2019;12(4):1272–1283.
148. Cheng G, Song Z, Liu Y, et al. Long noncoding RNA SNHG12 indicates the prognosis of prostate cancer and accelerates tumorigenesis via sponging miR-133b. J Cell Physiol. 2020;235(2):1235–1246. doi:10.1002/jcp.29039
149. Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. doi:10.18632/oncotarget.9350
150. Zhao HF, Zhang ZC, Shi BK, Jiang XZ. DANCR sponges miR-135a to regulate paclitaxel sensitivity in prostate cancer. Eur Rev Med Pharmacol Sci. 2019;23(16):6849–6857. doi:10.26355/eurrev_201908_18724
151. Sun B, Ke KB, Liu DF, et al. Long noncoding RNA SNHG14 acts as an oncogene in prostate cancer via targeting miR-613. Eur Rev Med Pharmacol Sci. 2020;24(2):633–638. doi:10.26355/eurrev_202001_20039
152. Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. LncRNA SNHG15 acts as an oncogene in prostate cancer by regulating miR-338-3p/FKBP1A axis. Gene. 2019;705:44–50. doi:10.1016/j.gene.2019.04.033
153. Wu X, Xiao Y, Zhou Y, Zhou Z, Yan W. lncRNA SNHG20 promotes prostate cancer migration and invasion via targeting the miR-6516-5p/SCGB2A1 axis. Am J Transl Res. 2019;11(8):5162–5169.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.