Back to Journals » International Journal of Nanomedicine » Volume 18
Long Non-Coding RNAs Within Macrophage-Derived Exosomes Promote BMSC Osteogenesis in a Bone Fracture Rat Model
Authors Wang D, Liu Y, Diao S, Shan L, Zhou J
Received 25 November 2022
Accepted for publication 17 February 2023
Published 28 February 2023 Volume 2023:18 Pages 1063—1083
DOI https://doi.org/10.2147/IJN.S398446
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Dong Wang, Yang Liu, Shuo Diao, Lei Shan, Junlin Zhou
Department of Orthopedics, Beijing Chaoyang Hospital, Capital Medical University, Beijing, 100020, People’s Republic of China
Correspondence: Junlin Zhou, Department of Orthopedics, Beijing Chaoyang hospital, Capital Medical University, 8 Gongren Tiyuchang Nanlu, Chaoyang District, Beijing, 100020, People’s Republic of China, Tel +86-13240718193, Email [email protected]
Purpose: To investigate the effect of macrophage exosomal long non-coding (lnc)RNAs on bone mesenchymal stem cell (BMSC) osteogenesis and the associated mechanism.
Methods: Rat BMSCs and spleen macrophages were co-cultured with serum derived from the fracture microenvironment of rat tibia. BMSC osteogenesis was evaluated using Alizarin red staining and the expression of BMP-2, RUNX2, OPN, and OC mRNA. BMSC osteogenesis was evaluated after co-culture with macrophages stimulated using hypoxic conditions or colony-stimulating factor (CSF). The uptake of macrophage-derived exosomes by BMSCs was evaluated using the exosome uptake assay. High-throughput sequencing and bioinformatics analyses were performed to identify key lncRNAs in the macrophage exosomes. The effect of lncRNA expression levels on BMSC osteogenesis was also assessed using a lncRNA overexpression plasmid and siRNA technology. M1 and M2 macrophages were distinguished using flow cytometry and the key exosomal lncRNA was detected by in situ hybridization.
Results: In the fracture microenvironment, macrophages (stimulated using either hypoxia or CSF) significantly increased the osteogenic ability of BMSCs. We showed that BMSCs assimilated macrophage-derived vesicles and that the inhibition of exosomal secretion significantly attenuated the macrophage-mediated induction of BMSC osteogenesis. The hypoxia condition led to the up-regulation of 310 lncRNAs and the down-regulation of 575 lncRNAs in macrophage exosomes, while CSF stimulation caused the up-regulation of 557 lncRNAs and the down-regulation of 407 lncRNAs. In total, 108 lncRNAs were co-up-regulated and 326 lncRNAs were co-down-regulated under both conditions. We eventually identified LOC103691165 as a key lncRNA that promoted BMSC osteogenesis and was expressed at similar levels in both M1 and M2 macrophages.
Conclusion: In the fracture microenvironment, M1 and M2 macrophages promoted BMSC osteogenesis by secreting exosomes containing LOC103691165.
Keywords: macrophage exosomes, BMSCs osteogenesis, lncRNAs, LOC103691165, bone fracture microenvironment
Introduction
Bone fracture is one of the most common clinical injuries and causes of human disability.1–3 Although fractures can be reduced and fixed, 5–10% of patients suffer from delayed fracture healing or nonunion.3–6 Therefore, clinical and basic medical research strive to promote fracture healing, reduce nonunion rate, and accelerate the rehabilitation of patients.
Bone mesenchymal stem cells (BMSCs) are the main source of osteoblasts in vivo; fracture healing is closely related to the osteogenic potential of BMSCs.7–10 Following a fracture, local and recruited BMSCs proliferate and differentiate into osteoblasts to initiate bone repair.9,10 Consequently, BMSCs are often used in tissue engineering to promote bone healing by implantation of materials carrying them or by direct cell injection.2,11,12 However, their curative effect is not stable and the BMSC-mediated regulation of osteogenesis is not completely understood.
Macrophages are one of the earliest cells to reach the fracture site. They secrete a variety of substances to regulate the inflammatory response and immune balance. In addition, macrophages recruit BMSCs to the site of fracture and promote their differentiation.7,13,14 Exosomes are a type of extracellular vesicle, 40–100 nm in diameter, actively secreted by cells.6,15,16 Exosome secretion is an important aspect of the macrophage regulatory mechanism.15–18 Inhibiting this process results in the loss of several macrophage functions.6,15–18 Treating macrophages with a sphingomyelinase inhibitor (GW4869), a widely recognized inhibitor of exosome secretion, dysregulates inflammatory homeostasis, immune balance, and the functions of cells such as cardiomyocytes and tumor cells.19,20
To date, several studies have explored the regulatory effect of macrophage exosomes on BMSC osteogenesis using mouse macrophage-like cell lines (RAW 264.7 cells, tumor cells) or stem cells stimulated with macrophage colony stimulating factor (CSF).21–24 However, these models may not accurately mimic the regulatory mechanisms of normal macrophages. In addition, the cells used in these studies were cultured in osteogenic induction medium, which may not adequately replicate the bone fracture microenvironment.21–24
Exosomes contain cellular substances such as proteins, lipids, and nucleic acids.25 Long non-coding RNAs (lncRNAs), which are RNAs with lengths greater than 200 nucleotides, are often found in exosomes, where they play important roles in exosomal function.26,27 Mi et al and Yin et al found that the down-regulation of the lncRNA FAP1-AS1 in macrophage exosomes reversed their ability to regulate tumor cell proliferation and migration.26,27 However, the effect of exosomal lncRNA on BMSC osteogenesis is still unknown.
In this study, we explored the role of macrophage exosomal lncRNA in BMSC osteogenesis. First, rat BMSCs and macrophages were extracted and identified. Second, the role of macrophages in BMSC osteogenesis was analyzed using a co-culture of BMSCs and macrophages. Third, a fluorescence tracer was used to explore whether macrophages secreted extracellular vesicles that affected BMSC function. In addition, the effect of exosomes on the macrophage-mediated induction of BMSC osteogenesis was clarified by using GW4869, which inhibited exosomal secretion in the bone fracture microenvironment. Fourth, key exosomal lncRNAs were identified using high-throughput sequencing and bioinformatics analysis. Finally, the regulatory effect of these key lncRNAs on BMSC osteogenesis was evaluated by altering their levels with an overexpression plasmid or silencing RNA technology.
In our study, hypoxia and CSF were used to stimulate macrophages. Sabi et al found that hypoxia plays an important role in the development of rheumatoid arthritis, possibly by promoting the synthesis and secretion of vascular endothelial growth factor in cells implicated in angiogenesis.28 Moreover, Behl et al found that hypoxia was closely related to the inflammatory response and MAPK signaling in rheumatoid arthritis.29 These results prompted us to use hypoxia and CSF to investigate the mechanism of macrophage-mediated regulation of BMSC osteogenesis.
Materials and Methods
Animals
Eighty female adult Sprague-Dawley (SD) rats (Vital River Laboratories, Laboratory Animal Institution Accreditation Certificate Registration Number: CNAS LA0004, 180–200 g) were kept in separate sterile cages and free access to sterilized chow diet and water. The feeding environment was specific pathogen free under a 12-h light/dark cycle at 23.6°C and 35% humidity. Animal welfare and experimental procedures were complied with the principles of Laboratory animal – Guidelines for ethical review of animal welfare (GB/T 35892–2018). Experimental protocols in this study were approved by the Capital Medical University ethics committee on the use of animals in research and education. The following humane endpoints were applied: extreme difficulty in moving to get food or water, severe wound infection, limb necrosis, automutilation, and screaming in response to a gentle touch.6,30
At the end of the experiments or when the humane endpoints were reached, the rats were euthanized via an excessive dose of sodium pentobarbital (100 mg/kg, intraperitoneal injection). The ethical code of animal experiments is AEEI-2022-290. Animal death was confirmed by the absence of breathing or heartbeat.
Reagents
All the reagents used are presented in Supplementary Material.
Isolation of BMSCs
BMSCs were isolated from ten rats, as previously reported.31 Briefly, the rats were sacrificed and their femurs and tibias were obtained aseptically. The bone marrow tissues were flushed and filtered through 200-mesh sieves to yield a single-cell suspension. Then, the cells were incubated in low-sugar Dulbecco’s Modified Eagle Medium (L-DMEM) supplemented with 10% fetal bovine serum (FBS), 1% streptomycin, and 1% penicillin. The cell culture medium was replaced with fresh medium every 3 days. The cells were used in experiments until they reached passage 5. BMSCs were identified using a combination of microscopy and flow cytometry.
Osteogenic, Adipogenic, and Chondrogenic Differentiation
The osteogenic, adipogenic, and chondrogenic induction media were purchased from Procell Life Science & Technology Co., Ltd. The osteogenic induction medium was composed of basal medium containing 10% FBS, 1% glutamine, 1% streptomycin, 1% penicillin, 1% β-glycerophosphate, 0.2% ascorbate acid, and 0.01% dexamethasone. The chondrogenic induction medium was composed of basal medium containing 0.1% sodium pyruvate, 1% ITS supplement, 1% TGF-β3, 0.3% ascorbic acid, 0.1% proline, 0.01% dexamethasone, 1% streptomycin, and 1% penicillin. Cells were cultured in differentiation induction medium for 14 days. Alizarin red and Alcian blue staining were used to verify the cell differentiation results.
Two types of adipogenic medium were used. The first medium type contained 10% FBS, 1% glutamine, 1% streptomycin, 1% penicillin, 0.2% insulin, 0.1% IBMX, 0.1% rosiglitazone, and 0.1% dexamethasone. The second medium type contained 10% FBS, 1% glutamine, 1% streptomycin, 1% penicillin, and 0.2% insulin. Cells were cultured in first medium type for 3 days and the second medium type for 1 day every 4 days until 16 days had passed. Oil Red O staining was used to verify the cell differentiation results.
Extraction of Macrophages
Sixty rats were used to extract macrophages; ten of these rats were also used to simultaneously extract the BMSCs. The method used was similar to that of previous studies.32 Briefly, spleens were obtained aseptically and ground in 4 °C precooled phosphate buffered saline (PBS), following by filtration through 200-mesh sieves to yield a single-cell suspension. The cells were then harvested and resuspended in L-DMEM with 10% FBS. After a 2 h incubation, the adherent cells were harvested. Cells were identified by flow cytometry.
Flow Cytometric Analysis
Cells were incubated with the following specific antibodies in the dark at 4 °C for 30 min: rabbit anti-CD29/Alexa Fluor 488 (1:100 dilution), rabbit anti-CD90/Alexa Fluor 488 (1:100 dilution), rabbit anti-CD44/Alexa Fluor 488 (1:100 dilution), rabbit anti-CD34/Alexa Fluor 488 (1:100 dilution), and rabbit anti-CD68/Alexa Fluor 488 (1:100 dilution) (all purchased from Bioss, China). The rabbit IgG/Alexa Fluor 488 (1:100 dilution; Bioss, China) was the isotype control antibody. Flow cytometry was used to detect the ratios of positively stained cells.
Alizarin Red Staining
The Alizarin red S staining kit (C0148S, Beyotime, China) was used to stain cells. Briefly, cells were washed three times with DPBS and then fixed using a cell fixative for 30 min. Then, Alizarin red staining solution was used to stain the cells at room temperature for 30 min. Finally, red mineralized nodules were observed. Absorbance at 570 nm was also subsequently measured on a microplate reader.
Alcian Blue Staining
Alcian blue staining was used to observe chondrogenic differentiation. Briefly, cells were washed with DPBS three times, and then fixed for 30 min using 4% paraformaldehyde. Alcian blue staining solution was then used to stain the cells at room temperature for 30 min. Finally, cells were observed on a microscope.
Oil Red O Staining
Lipid droplet formation was detected using oil red O staining. Briefly, cells were washed three times with DPBS and fixed using 4% paraformaldehyde for 30 min at room temperature. Oil red O staining solution was then added to stain the cells for 0.5 h. Finally, the formation of lipid droplets was captured on a microscope.
Rat Tibia Fracture Model
The tibia fracture model was described previously.6,33 Briefly, 20 rats were anaesthetized under an oxygen flow rate of 600 mL/min with 3% isoflurane. The right lower limb was shaved, disinfected with 75% ethanol, and covered with a sterile sheet. A 5 mm incision was made at the median of the right lower limb to expose the tibia shaft and a bone saw was used to create a transverse fracture. The fracture ends were cauterized using an electric knife and the fracture was fixed with a 0.8-mm intramedullary Kirschner wire (K-wire, Zimmer, USA). The incision was closed with a 5–0 absorbable suture (Ethicon, USA). Finally, intramuscular injections of gentamicin (2 mg/day) were administered postoperatively for 3 days to prevent infection.
Construction of Bone Fracture Environment Medium (BFM)
Briefly, 7 days after the tibia fracture model was established in 20 rats, peripheral blood was collected from the rats and centrifuged at 3500 rpm for 15 min. The rat serum was harvested and heated at 56 °C for 30 min to inactivate the complement proteins. Then, 10% serum, 1% streptomycin, 1% penicillin, and 94% L-DMEM were added to produce the BFM.6
Cell Co-Culture Model
Transwell inserts (12-well plate, pore size 1.0 µm) were used. Briefly, BMSCs were incubated in the lower chamber with 1 mL BFM and macrophages were incubated in the upper chamber with 0.5 mL BFM.
Hypoxia Stimulation
The hypoxia stimulation method used in this study was similar to that previously published.34 Briefly, cells were cultured in BFM with 1% O2, 5% CO2, and 94% N2 for 4 h and 95% air, 5% CO2 for 16 h. The hypoxia (4 h)/reoxygenation (16 h) cycle was repeated a total of 2 cycles.
Stimulation with CSF
The CSF stimulation method used in this study was similar to that of previous reports.35 Briefly, the cells were cultured in BFM with 10 ng/mL CSF for 48 h.
Quantitative Real-Time (qRT)-PCR
The expression of lncRNAs was analyzed by qRT-PCR. Total RNA was extracted using the Tissue RNA Extraction Kit (G3640-50T, Servicebio). Then, the SweScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (G3337-100, Servicebio, China) was used for reverse transcription into cDNA, based on the manufacturer’s instructions. The qRT-PCR was performed using the 2× Universal Blue SYBR Green qPCR Master Mix (G3326-05, Servicebio, China), following the manufacturer’s instructions. Table 1 shows the sequences of the PCR primers used in this study, which were designed using primer 5.0 software. Relative lncRNA or mRNA levels were normalized to U6 RNA or actin mRNA using the 2−ΔΔCT method, respectively.
 |
Table 1 Primer Sequences Used in This Study |
Exosome Extraction
Macrophage exosomes were extracted using the Cell Supernatant Exosome Extraction Kit (EX0011, Solarbio, China), in accordance with the manufacturer’s instructions.
Transmission Electron Microscopy (TEM)
TEM was performed by Wuhan Servicebio Biotechnology Co., Ltd. Briefly, 20 μL of the exosomal suspension was applied to the copper grid coated in carbon film for 5 min. Then, 2% phosphotungstic acid was added onto the copper grid for 2 min of staining. The cuprum grids were observed on a TEM (HT-7800, Hitachi, Japan).
Nanoparticle Tracking Analysis (NTA)
NTA was performed by Wuhan Servicebio Biotechnology Co., Ltd. using ZetaView (Particle Metrix). Exosomal samples were diluted in PBS (1:5000 dilution) and their size and concentration were analyzed using NTA video capture at 1401 positions at room temperature. The acquisition conditions were set as follows: Scattering Intensity: 4.0, Minimum Brightness: 30, Maximum Brightness: 255, Minimum Area: 10, Maximum Area: 1000, Minimum Trace length: 15, Gain: 28.80, Shutter: 500.00, Laser Wavelength (nm): 520.00, Micrometer per Pixel: 0.713, and Reference Electrode Distance: 5.15. The videos were analyzed using ZetaView software version 8.05.14.
Exosome Uptake Assay
Transwell inserts (12-well plate, pore size 1.0 µm) were used. Macrophages were stained with DiD (D4019, Yuheng) and BMSCs were stained by DiO (D4007, Yuheng), in accordance with the manufacturer’s instructions. The cell co-culture model was then constructed, and the hypoxia or CSF stimulation conditions were applied. After incubating for 24 and 48 h, the lower chamber was observed on an inverted fluorescence microscope.
High Throughput Sequencing and Bioinformatics Analysis
High-throughput sequencing was used to characterize the lncRNAs that were involved in the hypoxia- and CSF-mediated stimulation of macrophages; the methods used were similar to those of a previous study.36 Briefly, the BMSCs were divided into three groups: hypoxia stimulation, CSF stimulation, and control group (BFM stimulation only). After incubation for 48 h, the cells were harvested and sent to Beijing Echo Biotech Co. Ltd., for high-throughput sequencing and analysis.
The criteria for differentially expressed lncRNAs (DELs) were false discovery rate (FDR) and adjusted p-value < 0.05 and a log2 fold-change (FC) ≥ 1. The DELs were analyzed in the NCBI database to exclude the RNAs that were not fully characterized as lncRNAs at present. The transcriptional abundance (TA) of DELs at the intersection between “hypoxia stimulation vs control group” and “CSF stimulation vs control group” were analyzed. The DELs with TAs less than 100 in the hypoxia stimulation group and the CSF stimulation group were excluded. The intersection DEL results were sorted according to the formula: (hypoxia stimulation group TA + CSF stimulation TA)/2. qRT-PCR was used to verify the sequencing results of the top three DELs. Expression plasmids and small interfering (si)RNAs of DEL candidates, which were encoded by a defined gene were unpatented, were then constructed by Beijing Yibiake Biotechnology Co., Ltd.
Plasmid Extraction
The Free Endotoxin Plasmid Extraction Maxi Kit (D1150-10T, Solarbio) was used to extract the expression plasmid, in accordance with the manufacturer’s instructions.
In situ Hybridization and Immunofluorescence
Macrophages were sent to Wuhan Servicebio Biotechnology Co., Ltd. for in situ hybridization and immunofluorescence detection. The lncRNA LOC103691165 probe was composed of a mixture of the following sequences: 5’-CATCTGCTTTTTCGATCCAAACTCC-3’, 5’-GAGTCTCAAAATGGTTCCTGGTCTTA-3’, 5’-GGACCTCGTAACTGAAATCAAGCCT-3’, 5’-CTTGAAATGGCAGTGTCATCAGGTT-3’, and 5’-TGATAGAGGGGCATCGTCTTGGTT-3’. CD206 was used as a surface marker of M2 macrophages and CD86 was used as a surface marker of M1 macrophages. Information on the antibodies used in this study is supplied in Supplementary Table 1.
Statistical Analysis
SPSS 25.0 software was used to analyze the data. The Shapiro–Wilk test was used to evaluate the normality of the data. If the data were normally distributed, they were presented as the mean ± standard deviation (SD); an independent sample t-test was used to compare two groups, whereas one-way ANOVA was used to compare multiple groups. If the data assumed a skewed distribution, they were presented as the median plus interquartile range (IQR); the Mann–Whitney U-test was used to analyze differences between two groups, while the Kruskal–Wallis H-test was used to compare more than two groups. The data were and compared using Chi-squared test or Fisher's exact test. A p-value < 0.05 was used to indicate a statistically significant difference.
Results
The Characteristics of Macrophages and BMSCs
The macrophages extracted from the primary rat spleen were spherical and uniform in size (Figure 1a). The macrophage-specific marker, CD68, was highly expressed on cell surface of 90.7% of these macrophages (Figure 1b).
The extracted primary rat BMSCs were cultured until the fifth generation of daughter cells; these BMSCs were long, spindle-shaped, and translucent (Figure 1c). Alizarin red, Alcian blue, and Oil red O staining showed that the cells had osteogenic, chondrogenic, and adipogenic differentiation ability (Figure 1d–f). BMSC-specific markers, such as CD29 (92.6%), CD90 (98.8%), and CD44 (99.3%), were expressed on the surface of these cells (Figure 1g–i). Although 82.1% of the cells were CD34+, a lot of this CD34 staining was non-specific (as shown by the CD34 isotype control), meaning that the BMSCs actually exhibited low CD34 expression (Figure 1j).
The Effect of Macrophages on BMSC Osteogenesis in the Bone Fracture Microenvironment
Alizarin red staining was used to observe BMSC osteogenesis. BMSCs and macrophages were co-cultured in BFM using a 12-well transwell chamber plate. BMSCs were placed in the lower chamber, while the upper chamber was either population with macrophages or left empty. In addition, the cells in the hypoxia group were stimulated using hypoxic conditions (two cycles of culture in 1% O2, 5% CO2, and 94% N2 for 4 h and 95% air, 5% CO2 for 16 h) and the cells in the CSF group were stimulated using CSF (ie, by culturing with 10 ng/mL CSF for 48 h).
The results showed that BMSCs co-cultured with macrophages had better osteogenic ability. Observation under the microscope showed that macrophages increased the BMSC area stained by Alizarin red, regardless of whether they were stimulated by hypoxia, CSF, or BFM alone (Figure 2).
At the same time, we measured the absorbance of each cell culture well at 570 nm. We found that macrophages significantly increased the OD of Alizarin-red-stained BMSCs (p < 0.05).
In addition, we detected the expression of osteogenesis-related genes, such as BMP-2, RUNX2, OPN, and OC, which was significantly induced by macrophages of each BMSC treatment group (p < 0.05, Figure 3).
The Characteristics of Macrophage Exosomes and Their Assimilation by BMSCs
Exosomes are one of the important ways for macrophages to regulate cells. Thus, we further analyzed the role of exosomes in the macrophage-mediated induction of BMSC osteogenesis. First, we extracted exosomes secreted by macrophages into the fracture microenvironment. The morphology of exosomes was assessed using TEM. In the TEM images, the exosomes were spherical and uniform in size (Figure 4a). NTA was used to analyze exosomal size, which showed that exosomes were 100–200 nm in diameter, and were present at a concentration of 6.6×1010 particles/mL (Figure 4b and c). At the same time, we used Western blotting to detect the expression of Alix, CD63, TSG101, CD9, and GAPDH proteins on the extracted exosomes. The results showed that the exosomes expressed Alix, CD63, TSG101, and CD9, but not GAPDH (Supplementary Figures 1–5).
The exosome uptake assay was used to evaluate whether BMSCs assimilated exosomes secreted by macrophages. To this end, macrophages were labeled with red fluorescence and BMSCs were labeled with green fluorescence. A cell co-culture system was constructed using a transwell plate, whereby BMSCs were placed in the lower chamber. The results showed that at the end of the assay period, BMSCs contained the red fluorescence signals of macrophages, regardless of whether these macrophages were stimulated using hypoxia, CSF stimulation, or BFM alone. These data suggested that BMSCs absorbed extracellular vesicles secreted by macrophages (Figure 4d).
The Effect of Macrophage Exosomes on BMSC Osteogenesis in the Bone Fracture Microenvironment
We evaluated whether macrophages affected the osteogenic function of BMSCs by secreting exosomes. GW4869 was used to inhibit the secretion of exosomes by macrophages. As before, transwell plates were used to generate a cell co-culture system to replicate the fracture microenvironment. The osteogenic function of BMSCs was evaluated using Alizarin red staining. The results showed that inhibiting the secretion of exosomes by macrophages reduced the area Alizarin-red-stained BMSCs in the cell co-culture model (Figure 5). This suggested that inhibiting the secretion of exosomes by macrophages weakened the role of macrophages in promoting BMSCs osteogenesis, irrespectively of whether the macrophages were stimulated using hypoxia, CSF, or BFM alone.
Moreover, we detected the absorbance of BMSCs at 570 nm after Alizarin red staining. The results showed that inhibiting the secretion of exosomes by macrophages significantly reduced the absorbance value of stained BMSCs in the cell co-culture model (p < 0.05, Figure 5).
In addition, qRT-PCR was used to detect the expression genes associated with osteogenesis in BMSCs, namely BMP-2, RUNX2, OPN, and OC. The results showed that the inhibition of macrophage exosome secretion significantly reduced the mRNAs levels of these genes (p < 0.05, Figure 6).
Identification of DELs in Macrophage-Derived Exosomes
The previous results had shown that macrophages played an important role in BMSC osteogenesis by secreting exosomes. We performed high-throughput sequencing of lncRNAs in the macrophage-derived exosomes to explore the key lncRNAs that affected the osteogenesis of BMSCs. We extracted exosomes secreted by macrophages cultures under conditions of hypoxia, CSF stimulation, or BFM alone. The results showed that hypoxia in the fracture microenvironment caused the differential expression of 885 lncRNAs in macrophage exosomes; among them, 575 lncRNAs were down-regulated and 310 lncRNAs were up-regulated (Figure 7). CSF in the fracture microenvironment caused the differential expression of 964 lncRNAs in macrophage exosomes; among them, 557 lncRNAs were up-regulated and 407 lncRNAs were down-regulated (Figure 7). A Venn diagram was used to find the co-up-regulated lncRNAs and co-down-regulated lncRNAs in fracture microenvironment under hypoxic of CSF stimulation conditions. The results showed that 108 lncRNAs were co-up-regulated and 326 lncRNAs were co-down-regulated (Figure 7). TA was used to evaluate the content of lncRNAs in exosomes. Co-up/down-regulated DELs with TAs ≥ 100 were further analyzed, which revealed three lncRNAs: LOC102555570, LOC103691165, and LOC100909675 (Figure 7).
The Effect of Exosomal lncRNA LOC103691165 on BMSC Osteogenesis in the Bone Fracture Microenvironment
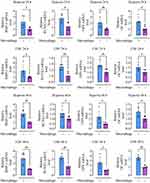
qRT-PCR was used to validate the sequencing results of LOC102555570, LOC103691165, and LOC100909675. The results showed that after the macrophages were stimulated using hypoxia or CSF, the above three lncRNAs were significantly up-regulated in the macrophage exosomes (p < 0.05, Figure 8).
The gene sequence expressing LOC102555570 was not clearly defined in the NCBI database, making it difficult to construct an overexpression plasmid. LOC100909675 was protected by patents and therefore could not be used in our research on tissue healing. Therefore, we constructed LOC103691165-overexpression plasmid and the corresponding siRNA. At the same time, exosomes secreted by macrophages in the fracture microenvironment were collected. Macrophage exosomes with upregulated, silenced, or normal LOC103691165 expression were produced using liposome transfection. The results showed that LOC103691165 overexpression increased the area of Alizarin-red-stained BMSCs, while LOC103691165 knockdown reduced the area of staining (Figure 8). In addition, we found that LOC103691165 overexpression significantly increased the absorbance of Alizarin-red-stained BMSCs at 570 nm, while LOC103691165 knockdown significantly decreased the absorbance (p < 0.05 for both, Figure 8).
When the expression of genes closely related to osteogenesis in BMSCs was examined, we found that LOC103691165 overexpression significantly promoted the expression of BMP-2, RUNX2, OPN and OC, while LOC103691165 silencing significantly inhibited their expression (p < 0.05 for both, Figure 8).
The Source of Macrophage-Derived Exosomal lncRNA LOC103691165 in the Bone Fracture Microenvironment
The fluorescent probe was used to determine which macrophages expressed LOC103691165 by in situ hybridization. CD86 was used to label M1 macrophages, and CD206 was used to label M2 macrophages. Macrophages were obtained after stimulation with hypoxia, CSF, or BFM alone. Fluorescence results showed that both the M1 and M2 macrophages expressed LOC103691165 (Figure 9). Cells were counted using DAPI and five viewing fields were randomly selected. The results showed that almost all M1 and M2 macrophage expressed LOC103691165, and that there was no significant difference in the LOC103691165 expression between them (p > 0.05, Figure 9).
Discussion
MSCs are the main source of osteoblasts in vivo, and their osteogenic ability is closely related to fracture healing.37,38 BMSCs are often implanted on scaffolds or directly injected into the site of the fracture to promote bone healing.39 However, the regulatory mechanism of BMSC osteogenesis is not fully understood.2,11,12
Macrophages are one of the earliest cells to reach the fracture site. On doing so, they secrete a variety of substances that regulate the inflammatory response and immune homeostasis, and promote cell differentiation.14 Secretion of exosomes is one of the key ways that macrophages play a regulatory role. LncRNAs are an important component of exosomes and play an important role in exosomal function.26,27 At present, the effect of macrophage-derived exosomal lncRNAs on BMSC osteogenesis in the fracture microenvironment is unclear.
In this study, we extracted and identified rat BMSCs and macrophages. In addition, the fracture microenvironment was reconstituted in vitro according to previously published methods.6 We then explored the mechanism of the macrophage-mediated regulation of BMSC osteogenesis in the fracture microenvironment. We selected the two most commonly used methods for stimulating macrophages and identified exosomal DELs at the intersection between both conditions. Through the logic of covariation, we were able to clarify the mechanism of macrophage-mediated regulation of BMSCs.
First, we analyzed whether macrophages regulated BMSC osteogenesis. Although such regulation has been confirmed by many studies, Li et al23 found that exosomes derived from M2 macrophages inhibited adipogenesis and promoted osteogenesis by BMSCs. In addition, treatment of BMSCs with M2 macrophage derived exosomes increased the expression of miR-690, IRS-1, and TAZ in BMSCs. Xiong et al24 found that miR-5106 expression was significantly higher in M2 macrophage exosomes and lower in M1 macrophage exosomes. In addition, miR-5106 promoted BMSC osteogenic differentiation. However, these studies did not analyze cells in the fracture microenvironment or use normal rat-derived macrophages. The osteogenic ability of BMSCs was evaluated by Alizarin red staining and osteogenic gene expression. We found that hypoxia and CSF both promoted BMSC osteogenesis in the fracture microenvironment. Macrophages further enhanced this osteogenic effect. Previously, there has been no study of the macrophage-mediated regulation of BMSC osteogenesis in the fracture microenvironment under hypoxic conditions or when stimulated by CSF.
We analyzed regulatory mechanisms by extracting macrophage exosomes in the fracture microenvironment and identifying them by TEM, NTA, and Western blotting. We co-cultured cells in the fracture microenvironment and found that macrophages secreted exosomes that acted on BMSCs. Prolonging co-culture time increased the number of macrophage exosomes acting on BMSCs. Since the macrophages secreted BMSC-modulating exosomes, we next explored whether BMSC osteogenesis was affected by these exosomes. GW4869 was used to inhibit the secretion of exosomes, which reduced BMSC osteogenesis, suggesting their importance in this process. Previous studies that have demonstrated this relationship used tumor-derived macrophages (RAW 264.7 cells) or macrophages transformed from stem cells.21–24 The results obtained using these types of macrophages may not be representative of the macrophages found in normal tissues. Moreover, most of these studies were carried out in osteogenic induction medium, which may not mimic the fracture microenvironment.21–24 Osteogenic induction medium also interferes with the osteogenesis of BMSCs, making it more difficult to judge whether macrophages or the culture medium promote BMSC osteogenesis.
LncRNAs are key components of exosomes and play important roles in exosomal function. There have been no previous studies evaluating the effect of lncRNAs originating from macrophage exosomes on BMSC osteogenesis. We used high-throughput sequencing to analyze DELs in macrophage exosomes in response to hypoxic conditions or CSF stimulation. Then, through the principle of covariation, the intersecting DELs were selected. Based on their TA, we identified LOC102555570, LOC103691165, and LOC100909675 as potentially the most important lncRNAs in macrophage exosomes involved in BMSC-mediated osteogenesis.
qRT-PCR validated the high-throughput sequencing results. Hypoxia or CSF significantly up-regulated the expression of LOC102555570, LOC103691165, and LOC100909675 in macrophage exosomes. However, the sequence of the gene encoding LOC102555570 was unclear, and an overexpression plasmid was difficult to construct. In addition, LOC100909675 was protected by a patent, meaning that it could not be used in our research. Thus, we chose LOC103691165 for further study.
There have been no studies of LOC103691165 to date, so the present study was the first to explore its function. We found that the up-regulation of LOC103691165 in macrophage exosomes promoted BMSC osteogenesis, indicating that LOC103691165 was closely implicated in this process. We subsequently evaluated whether M1 or M2 macrophages were the source of LOC103691165. Surprisingly, both M1 and M2 macrophages synthesized LOC103691165, and there was no significant difference in the expression levels between the two. These results show that both M1 and M2 macrophages may promote BMSCs osteogenesis.
At present, the question of whether M1 or M2 macrophages promote BMSC osteogenesis remains controversial. Some studies have suggested that M2 macrophages promote BMSC osteogenesis, while M1 macrophages have an inhibitory effect.21,24 However, other studies came to different conclusions, proposing that exosomes from M1 macrophages promoted osteogenesis.40 Of note, the source of macrophages used in these studies was either RAW 264.7 cells or CSF-transformed stem cells, which may have generated inaccurate results.
In our study, BMSC mineralization occurred within 48 h, which was much faster than is achieved using traditional osteogenic stimulation. This result might be explained by the fact that in our study we used the fracture microenvironment and macrophage co-culture and applied hypoxia or CSF stimulation at levels greater than those used in traditional osteogenic induction. Moreover, BMSC mineralization was also reported to occur within 48 h by many other studies, which was consistent with our observations.41,42 However, elucidating the specific mechanism will require further study.
An important goal of the approach used here was to analyze the effect of macrophages on BMSC osteogenesis and explore its mechanism in a fracture microenvironment. Macrophage exosomes were extracted from the fracture microenvironment and were then subjected to high-throughput sequencing to identify key lncRNAs. We found that LOC103691165 was present in the exosomes of both M1 and M2 macrophages and promoted BMSC osteogenesis.
There were several limitations to this study. First, our in vitro experiments simulated but did not exactly replication the in vivo fracture microenvironment, which will certainly be more complex. Second, we used transwell plates to construct a cell co-culture model, which prevented direct contact between cells. Although this simplified the study of the role of macrophage exosomes, it could not fully mimic the interaction between BMSCs and macrophages in vivo, as would involve direct cell-cell contact. Third, we identified the important lncRNAs in macrophage exosomes by high-throughput sequencing, bioinformatics analysis, and qRT-PCR. The functional analysis showed that LOC103691165 promoted BMSC osteogenesis. However, whether LOC103691165 is the most important lncRNA in macrophage exosomes is unknown. At present, the expression gene sequences of many lncRNAs are still not fully characterized, meaning that it is not always clear whether a given RNA is a lncRNAs, an mRNAs, or a meaningless RNA fragment. Finally, the mechanism of the LOC103691165-mediated regulation of BMSC osteogenesis, including the possible involvement of downstream proteins, has not been studied. These questions require further research.
In the future, we will further explore the mechanism by which macrophage exosome lncRNA LOC103691165 promotes BMSC osteogenesis and consider its activity in the context of clinical practice. We will analyze proteins that may bind to LOC103691165 using RNA pull-down experiments and mass spectrometry. In addition, we will study the microRNAs regulated by LOC103691165 using a luciferase reporter assay and explore its binding to these microRNAs and to proteins.
Conclusion
To the best of our knowledge, this study was the first to demonstrate that macrophages promoted BMSC osteogenesis by secreting exosomes into the fracture microenvironment. We found that exosomal lncRNA LOC103691165, expressed by both M1 and M2 macrophages, played an important role in this process. However, the fracture microenvironment in vivo is likely to be far more complex than our in vitro simulation and we did not factor in the impact of direct cell-cell contact. In the future, we will further explore the mechanism by which macrophage exosomal lncRNA LOC103691165 promotes BMSC osteogenesis and consider its activity in the context of clinical practice.
Abbreviations
BMSCs, Bone mesenchymal stem cells; CSF, macrophage colony stimulating factor; lncRNAs, Long non-coding RNAs; SD, Sprague-Dawley; L-DMEM, Dulbecco’s modified Eagle’s medium with low sugar; FBS, fetal bovine serum; BFM, bone fracture environment; TEM, transmission electron microscopy; NTA, nanoparticle tracking analysis; DELs, different expressed lncRNAs; FDR, false discovery rates; TA, transcriptional abundance; siRNA, silence RNA.
Data Sharing Statement
The Supplemental Data used to support the findings of this study have been submitted alongside this article.
Ethics Approval
This article does not contain any studies with human participants performed by any of the authors. The present study was approved by the Capital Medical University Ethics Committee on the use of animals in research and education (approval no. AEEI-2022-290).
Acknowledgments
We thank the Beijing Chaoyang hospital and the Capital Medical University for providing the laboratory space and all software used in this study. We thank staff at the Wuhan Servicebio Technology Co., Ltd for qRT-PCR primer construction, TEM, NTA, in situ hybridization, and immunofluorescence experiments and analyses. Finally, we thank Beijing Echo Biotech Co. Ltd. for the high-throughput sequencing and analysis.
Author Contributions
All authors have made a significant contribution to the work reported, be it in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all of these areas. All authors also took part in the drafting, revision, or critical review of the manuscript, have approved the version submitted for publication, and have agreed on the journal to which the manuscript has been submitted. The authors agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 82272469) and the Beijing Key Clinical Specialty Project.
Disclosure
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
1. Wildemann B, Ignatius A, Leung F, et al. Non-union bone fractures. Nat Rev Dis Primers. 2021;7(1):57. doi:10.1038/s41572-021-00289-8
2. Toosi S, Behravan N, Behravan J. Nonunion fractures, mesenchymal stem cells and bone tissue engineering. J Biomed Mater Res A. 2018;106(9):2552–2562. doi:10.1002/jbm.a.36433
3. Nicholson JA, Makaram N, Simpson A, Keating JF. Fracture nonunion in long bones: a literature review of risk factors and surgical management. Injury. 2021;52(Suppl 2):S3–S11. doi:10.1016/j.injury.2020.11.029
4. Ho-Shui-Ling A, Bolander J, Rustom LE, et al. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi:10.1016/j.biomaterials.2018.07.017
5. Nauth A, Lee M, Gardner MJ, et al. Principles of nonunion management: state of the art. J Orthop Trauma. 2018;32(Suppl 1):S52–S57. doi:10.1097/BOT.0000000000001122
6. Wang D, Wang J, Zhou J, Zheng X. The role of adenosine receptor A2A in the regulation of macrophage exosomes and vascular endothelial cells during bone healing. J Inflamm Res. 2021;14:4001–4017. doi:10.2147/JIR.S324232
7. Pajarinen J, Lin T, Gibon E, et al. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials. 2019;196:80–89. doi:10.1016/j.biomaterials.2017.12.025
8. Bahney CS, Zondervan RL, Allison P, et al. Cellular biology of fracture healing. J Orthop Res. 2019;37(1):35–50. doi:10.1002/jor.24170
9. Zhao Q, Liu X, Yu C, Xiao Y. Macrophages and bone marrow-derived mesenchymal stem cells work in concert to promote fracture healing: a brief review. DNA Cell Biol. 2022;41(3):276–284. doi:10.1089/dna.2021.0869
10. Bragdon BC, Bahney CS. Origin of reparative stem cells in fracture healing. Curr Osteoporos Rep. 2018;16(4):490–503. doi:10.1007/s11914-018-0458-4
11. Lin H, Sohn J, Shen H, Langhans MT, Tuan RS. Bone marrow mesenchymal stem cells: aging and tissue engineering applications to enhance bone healing. Biomaterials. 2019;203:96–110. doi:10.1016/j.biomaterials.2018.06.026
12. Lopez CD, Bekisz JM, Corciulo C, et al. Local delivery of adenosine receptor agonists to promote bone regeneration and defect healing. Adv Drug Deliv Rev. 2019;146:240–247. doi:10.1016/j.addr.2018.06.010
13. Yahara Y, Ma X, Gracia L, Alman BA. Monocyte/macrophage lineage cells from fetal erythromyeloid progenitors orchestrate bone remodeling and repair. Front Cell Dev Biol. 2021;9:622035. doi:10.3389/fcell.2021.622035
14. Schlundt C, Fischer H, Bucher CH, et al. The multifaceted roles of macrophages in bone regeneration: a story of polarization, activation and time. Acta Biomater. 2021;133:46–57. doi:10.1016/j.actbio.2021.04.052
15. Lu J, Wang QY, Sheng JG. Exosomes in the repair of bone defects: next-generation therapeutic tools for the treatment of nonunion. Biomed Res Int. 2019;2019:1983131. doi:10.1155/2019/1983131
16. Bei HP, Hung PM, Yeung HL, Wang S, Zhao X. Bone-A-Petite: engineering exosomes towards bone, osteochondral, and cartilage repair. Small. 2021;17(50):e2101741. doi:10.1002/smll.202101741
17. Zhang L, Jiao G, Ren S, et al. Exosomes from bone marrow mesenchymal stem cells enhance fracture healing through the promotion of osteogenesis and angiogenesis in a rat model of nonunion. Stem Cell Res Ther. 2020;11(1):38. doi:10.1186/s13287-020-1562-9
18. Liu W, Li L, Rong Y, et al. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020;103:196–212. doi:10.1016/j.actbio.2019.12.020
19. Wang Y, Qiu Z, Yuan J, et al. Hypoxia-reoxygenation induces macrophage polarization and causes the release of exosomal miR-29a to mediate cardiomyocyte pyroptosis. Vitro Cell Dev Biol Anim. 2021;57(1):30–41. doi:10.1007/s11626-020-00524-8
20. Zheng P, Luo Q, Wang W, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9(4):434. doi:10.1038/s41419-018-0465-5
21. Kang M, Huang CC, Lu Y, et al. Bone regeneration is mediated by macrophage extracellular vesicles. Bone. 2020;141:115627. doi:10.1016/j.bone.2020.115627
22. Liu K, Luo X, Lv ZY, et al. Macrophage-derived exosomes promote bone mesenchymal stem cells towards osteoblastic fate through microRNA-21a-5p. Front Bioeng Biotechnol. 2022;9:801432. doi:10.3389/fbioe.2021.801432
23. Li Z, Wang Y, Li S, Li Y. Exosomes derived from M2 macrophages facilitate osteogenesis and reduce adipogenesis of BMSCs. Front Endocrinol. 2021;12:680328. doi:10.3389/fendo.2021.680328
24. Xiong Y, Chen L, Yan C, et al. M2 Macrophagy-derived exosomal miRNA-5106 induces bone mesenchymal stem cells towards osteoblastic fate by targeting salt-inducible kinase 2 and 3. J Nanobiotechnology. 2020;18(1):66. doi:10.1186/s12951-020-00622-5
25. Shan X, Zhang C, Mai C, et al. The biogenesis, biological functions, and applications of macrophage-derived exosomes. Front Mol Biosci. 2021;8:715461. doi:10.3389/fmolb.2021.715461
26. Mi X, Xu R, Hong S, et al. M2 macrophage-derived exosomal lncRNA AFAP1-AS1 and MicroRNA-26a affect cell migration and metastasis in esophageal cancer. Mol Ther Nucleic Acids. 2020;22:779–790. doi:10.1016/j.omtn.2020.09.035
27. Yin Z, Zhou Y, Ma T, et al. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J Cell Mol Med. 2020;24(9):5028–5038. doi:10.1111/jcmm.15125
28. Sabi EM, Singh A, Althafar ZM, et al. Elucidating the role of hypoxia-inducible factor in rheumatoid arthritis. Inflammopharmacology. 2022;30:737–748. doi:10.1007/s10787-022-00974-4
29. Behl T, Upadhyay T, Singh S, et al. Polyphenols targeting MAPK mediated oxidative stress and inflammation in rheumatoid arthritis. Molecules. 2021;26:6570. doi:10.3390/molecules26216570
30. Zheng X, Wang J, Zhou J, Wang D. The extract of ilex cornuta bark promotes bone healing by activating adenosine A2A receptor. Drug Des Devel Ther. 2022;16:2569–2587. doi:10.2147/DDDT.S362238
31. Wang L, Li Y, Ren M, et al. pH and lipase-responsive nanocarrier-mediated dual drug delivery system to treat periodontitis in diabetic rats. Bioact Mater. 2022;18:254–266. doi:10.1016/j.bioactmat.2022.02.008
32. Xiang W, Shi R, Kang X, et al. Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat Commun. 2018;9(1):2574. doi:10.1038/s41467-018-04999-8
33. Zheng X, Wang D. The adenosine A2A receptor agonist accelerates bone healing and adjusts Treg/Th17 cell balance through interleukin 6. Biomed Res Int. 2020;2020:2603873. doi:10.1155/2020/2603873
34. Nieborowska-Skorska M, Kopinski PK, Ray R, et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood. 2012;119(18):4253–4263. doi:10.1182/blood-2011-10-385658
35. Zhang Y, Wang H, Zhu G, Qian A, Chen W. F2r negatively regulates osteoclastogenesis through inhibiting the Akt and NFκB signaling pathways. Int J Biol Sci. 2020;16(9):1629–1639. doi:10.7150/ijbs.41867
36. Wang D, Liu Y, Yang X, Zhou J. Hypoxic preconditioning enhances cell hypoxia tolerance and correlated lncRNA and mRNA analysis. Life Sci. 2018;208:46–54. doi:10.1016/j.lfs.2018.07.014
37. Shang F, Yu Y, Liu S, et al. Advancing application of mesenchymal stem cell-based bone tissue regeneration. Bioact Mater. 2020;6(3):666–683. doi:10.1016/j.bioactmat.2020.08.014
38. Fu X, Liu G, Halim A, et al. Mesenchymal stem cell migration and tissue repair. Cells. 2019;8(8):784. doi:10.3390/cells8080784
39. Tan SHS, Wong JRY, Sim SJY, et al. Mesenchymal stem cell exosomes in bone regenerative strategies-A systematic review of preclinical studies. Mater Today Bio. 2020;7:100067. doi:10.1016/j.mtbio.2020.100067
40. Xia Y, He XT, Xu XY, et al. Exosomes derived from M0, M1 and M2 macrophages exert distinct influences on the proliferation and differentiation of mesenchymal stem cells. Peer J. 2020;8:e8970. doi:10.7717/peerj.8970
41. Kim TY, Park JK, Prasad Aryal Y, et al. Facilitation of bone healing processes based on the developmental function of Meox2 in tooth loss lesion. Int J Mol Sci. 2020;21:8701. doi:10.3390/ijms21228701
42. Chen Z, Luo Q, Lin C, Kuang D, Song G. Simulated microgravity inhibits osteogenic differentiation of mesenchymal stem cells via depolymerizing F-actin to impede TAZ nuclear translocation. Sci Rep. 2016;6:30322. doi:10.1038/srep30322
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.