Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA LINC00662 Regulated Proliferation and Migration by Targeting miR-34a-5p/LMAN2L Axis in Glioma
Authors Geng Y , Wu Y, Xu C, Li T, Zhang L
Received 21 July 2020
Accepted for publication 4 September 2020
Published 9 October 2020 Volume 2020:13 Pages 10161—10172
DOI https://doi.org/10.2147/OTT.S272616
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Yibo Geng,1,* Yuliang Wu,2,* Cheng Xu,1 Tian Li,1 Liwei Zhang1,3
1Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Department of Neurosurgery, Qilu Children’s Hospital, Shandong University, Jinan, Shandong, People’s Republic of China; 3China National Clinical Research Center for Neurological Disease, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liwei Zhang
Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, No. 119 Nan Si Huan West Road, Fengtai District, Beijing 100070, People’s Republic of China
Tel +86-15101181174
Fax +86-010-59976611
Email [email protected]
Background: Numerous studies suggest that long non-coding RNAs (lncRNAs) participate in the biological process of diverse malignancies, including glioma. Although many differentially expressed lncRNAs have been identified in glioma, to our best knowledge, the role of LINC00662 and its potential underlying mechanism in glioma progression remains unclear. This study aimed to explore the function and regulatory network of LINC00662 in glioma.
Methods: Expressions of LINC00662, miR-34a-5p and lectin mannose-binding 2-like (LMAN2L) in glioma tissues were analyzed using The Cancer Genome Atlas Program (TCGA) and the Chinese Glioma Genome Atlas (CGGA) databases. Colony formation, Celltiter-Glo and BrdU (5-bromo-2ʹ-deoxyuridine) incorporation assays were used to detect cell proliferation in vitro. Xenograft mouse models were established to determine cell proliferation in vivo. Transwell and wound healing assay was used to detect cell migration. In addition, epithelial–mesenchymal transition (EMT) markers were detected by Western blot. Annexin V and 7-AAD were used to stain apoptotic cells. Interactions between miR-34a-5p and LINC00662 or the 3′-UTR of LMAN2L were predicted and determined by bioinformatics analysis, luciferase reporter assay and RNA immunoprecipitation (RIP) assays.
Results: High LINC00662 level predicted poor overall survival of glioma patients. Functional studies revealed that suppression of LINC00662 remarkably inhibited cell proliferation, clonogenicity and EMT pathway. Mechanistically, LINC00662 sponged miR-34a-5p to regulate LMAN2L expression. Furthermore, miR-34a-5p inhibitor reversed the anti-proliferation and anti-migration effect of LINC00662 knockdown, which could be rescued by downregulation of LMAN2L in glioma cells.
Conclusion: Our study was the first to report that LINC00662 acted as a competing endogenous RNA (ceRNA) to regulate glioma progression by targeting miR-34a-5p/LMAN2L axis, providing a new therapeutic target for glioma.
Keywords: long non-coding RNA, LINC00662, miR-34a-5p, LMAN2L, glioma, epithelial–mesenchymal transition
Introduction
Glioma is considered as the most lethal primary brain tumor, accounting for 80% of all central nervous system malignant tumors.1 Despite great efforts have dedicated to the therapeutic strategies for glioma over the past years, including surgery, radiotherapy and chemotherapy, the 5-year survival rate of glioma patients remains dismal.2 Thus, elucidating the molecular mechanisms underlying glioma and identifying effective biomarkers are essential to developing effective treatment modalities for this deadly malignancy.
The long non-coding RNAs (lncRNAs) are new-founded members which occupy a large portion of the non-coding RNA family.3 They are a class of transcripts with a length of more than 200 nucleotides, with rare protein-coding capacity.4 Accumulating evidence has confirmed that lncRNAs function as ceRNAs to regulate the expression of microRNAs (miRNAs), and further affect the target proteins.5 On the other hand, lncRNAs play a significant role in the regulation of cell proliferation, metastasis and EMT of various cancers.6 Specifically, LINC00662, which is located at the human chromosome 19q11,7 was originally identified as an oncogene in lung cancer.8 Subsequent studies suggested that LINC00662 was upregulated and promoted tumor progression in other malignancies, including gastric,9 oral,10 prostate,11 liver and colorectal cancer.12,13 The involvement of LINC00662 in glioma has not yet been documented, inspiring us to investigate its biological functions and the underlying mechanism.
In the current study, we analyzed that LINC00662 exacerbated glioma malignant phenotypes and predicted glioma patient survival outcomes. Xenograft mouse models were established to assess the tumor-promoting role of LINC00662 in vivo. Mechanistically, LINC00662 functioned as a ceRNA that upregulated LMAN2L by sponging miR-34a-5p, and their effects on glioma progression were also investigated.
Materials and Methods
Data Acquisition and Bioinformatics Analysis
The expression levels and survival risk of LINC00662, miR-34a-5p and predicted target genes (ARID4B, ATMIN, DAAM1, DCAF7, ELL2, FOXP1, GLCE, LMAN2L, METAP1, TMEM109, VAMP2 and VCL) were analyzed using gene expression profiling interactive analysis (GEPIA),14 TCGA or Chinese Glioma Genome Atlas (CGGA) database. The correlation expression between LINC00662, miR-34a-5p and predicted target genes were evaluated by The Encyclopedia of RNA Interactomes (ENCORI).15 ENCORI, Targetscan, miRDB and mirDIP were applied to predict the potential binding sites between LINC00662, miR-34a-5p and LMAN2L.16–18
Cell Culture
The human high-grade glioma cell lines, U87, U251, U343 and normal astrocyte cell line HEB were kindly gifts from Dr. Chen or purchased from Beijing winter song Boye Biotechnology (Beijing, China). A known problematic cell line U87 was authenticated by STR profile. The above-mentioned cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning, NY, USA) containing 10% fetal bovine serum (FBS) (HyClone, Logan, UT, USA) in a humidified environment at 37°C with 5% CO2. TT150630 and TT150714 were cultivated as previously described.19 All cell lines were confirmed to be negative for mycoplasma.
RNA Interference
Short-hairpin RNAs (shRNAs) against LINC00662 and LMAN2L were cloned in FUGW-H1 (H1) lentiviral plasmid as previously described and H1 was used as control.20 The lentivirus package was performed according to Hu’s protocol.21 miR-34a-5p inhibitor, mimics and their control miR-NC were obtained from GenePharm (Shanghai, China). For transfection, Lipofectamine 3000 (Invitrogen Life Technologies, Shanghai, China) was used according to the manufacturer’s guide. The RNA interference sequences are listed in Table 1.
 |
Table 1 RNA Interference Sequences |
RNA Isolation and qRT-PCR Analysis
The total RNA was extracted from the cell lines using TRIzol reagent (Thermo Fisher Scientific, MA, USA). qRT‐PCR was carried out in a Bio‐Rad CFX384 system (Bio-Rad, Hercules, CA, USA) and SYBR Green (CWBIO, Beijing, China) was utilized to evaluate the expression of LINC00662, LMAN2L and another eight predicted target genes. TaqMan MicroRNA Reverse Transcription kit and TaqMan Universal Master Mix II (Applied Biosystems, Foster City, CA, USA) were used to detect the expression levels of miR‐34a‐5p. Relative gene expression was figured out by 2-ΔΔCt method and normalized to GAPDH or U6. The primer sequences are provided in Table 2.
 |
Table 2 qRT-PCR Primer Sequences |
Cell Proliferation and Colony Formation Assay
The Celltiter-Glo kit (Promega, Madison, WI, USA) was used to detect cell viability and the luminescence intensity was evaluated by the TECAN Infinite 2000 plate reader (TECAN, Maennedorf, Zürich, Switzerland). To conduct colony formation assay, 1,000 cells/well were cultured in triplicates in 6-well plates and incubated at 37°C. Ten days later, the cells were fixed with methanol and stained with 0.5% crystal violet (Sangon Biotech).
Wound Healing
4×105 cells/well were seeded into 6-well plates and incubated for 24 hrs. Wounds were created by scratching the cell layer with a pipette tip. Then, the cells were cultured in the DMEM containing 2% FBS for another 24 hrs. Next, images were captured using a microscope (Zeiss, Jena, Germany).
BrdU Incorporation Assay
20 μM BrdU was applied to label cells for 15 minutes at 37°C. Next, cells were fixed by 4% paraformaldehyde (PFA) for 10 minutes, followed by denaturation with 2N HCl for 1hr and then neutralized with 20 mM Na2CO3 for another 1 hr. Subsequently, we blocked the cells with 5% bovine serum albumin and incubated the cells with anti-BrdU (Abcam) primary antibody overnight. DAPI (Sigma) was added during secondary antibody incubation. The signal was visualized using LSM880 confocal microscope (Zeiss).
Western Blot
The cell lysate preparation, membrane transfer and band visualization were performed using standard procedures. Primary antibodies (1:1000) used in our study listed as below: N-Cadherin (BD Biosciences, San Jose, CA, USA), Vimentin (Thermo Fisher Scientific), LMAN2L (Novus Biologicals, CO, USA) and GAPDH (abm, Zhenjiang, Jiangsu, China). The uncropped images of membranes used for our study were provided in Figure S1.
In vivo Experiments
All in vivo research was conducted strictly following the protocol of Care and Use of Laboratory Animals and approved by the Committee on the Ethics of Animal Experiments of Beijing Tiantan Hospital. 106 U87 cells stably expressing H1 or shLINC00662-1 were subcutaneously injected into the four-week-old female BALB/C nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China). Tumor growth rates were monitored at the indicated time. The tumor volume was calculated according to the formula: volume = 0.5 × length × width2. After four weeks, the mice were euthanized by CO2 and the tumors were weighed and fixed by 4% PFA.
For HE and immunohistochemistry staining, serial sections at 5 μm thickness were prepared and deparaffinized according to standard protocol. Next, the sections were stained with hematoxylin and eosin (Sangon Biotech) in accordance with manufacturer’s guideline. Other slides were incubated with anti-Ki67 primary antibody (1:1000, Invitrogen) overnight at 4°C. Subsequently, sections were visualized using DAB and co-stained with hematoxylin (Sangon Biotech), mounted with neutral gum and captured by the microscope (Zeiss).
Dual-Luciferase Reporter Assay
The wild type (WT) and mutant (MU) miR-34a-5p binding sites to LINC00662 sequence or LMAN2L 3′-UTR were separately cloned to pmirGLO (Promega) vectors to obtain LINC00662-WT/MU and LMAN2L-WT/MU vectors. The miR-34a-5p mimics, miR-NC were co-transfected into U87 and U251 cells with above luciferase vectors for 48 hours and finally examined using the Dual-Luciferase Assay System (Promega).
RNA Immunoprecipitation (RIP)
RIP assay was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Darmstadt, Germany) in accordance with the manufacturer’s protocol. Briefly, the cell extract was prepared and incubated with magnetic beads precoated with the Ago2-antibody or with the control IgG (Millipore). The precipitated complex was tested by qRT-PCR.
Statistics
Data were analyzed with GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) and were presented as mean ± standard deviation (SD). All the experiments were repeated three times at least. For the Western blot, representative images from three biological replicates were shown and the bands were analyzed with Image J and normalized to the loading controls. Kaplan–Meier method and Log rank test were carried out for survival analysis. The association of the expression of miR-34a-5p with LINC00662 and LMAN2L was analyzed by Pearson’s correlation. The comparison between the two groups was analyzed using a non-paired Student’s t-test. One-way ANOVA with Tukey’s post hoc test was used to test differences among multiple groups. P-value <0.05 was considered to indicate statistical significance.
Results
Upregulated LINC00662 Indicated Unsatisfactory Prognosis in Glioma
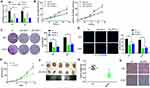
To determine the expression of LINC00662 and the relative prognosis in glioma, we used the data retrieved from the TCGA and CGGA platform. The results showed an increased expression trend of LINC00662 in glioma tissues compared with normal tissues (Figure 1A). Next, glioma patients were classified into high and low expression groups according to the median expression of LINC00662. Kaplan-Meier curve indicated that high LINC00662 expression displayed remarkably shorter overall survival in glioma patients (Figure 1B and C). Additionally, the expression level of LINC00662 in glioma cells was higher than normal astrocyte (Figure 1D). Both U87 and U251 cells were selected for further experiments due to their high LINC00662 expression levels.
Suppression of LINC00662 Inhibited Glioma Cell Proliferation Both in vitro and in vivo
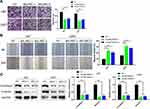
To evaluate the function of LINC00662 in glioma, we stably knocked down the expression of LINC00662 by three shRNA (shLINC00662-1, shLINC00662-2, shLINC00662-3) in U87 and U251 cells. As a result, shLINC00662-1 and shLINC00662-3 exhibited the most evident silencing effects and were selected for the subsequent experiments (Figure 2A). Celltiter-Glo assay indicated that LINC00662 knockdown markedly inhibited the proliferative capabilities of U87 and U251 cells (Figure 2B). The colony formation and BrdU incorporation assays were utilized to further confirm the inhibitory effects of LINC00662 knockdown on glioma cell proliferation (Figure 2C and D).
Since shLINC00662-1 showed better anti-tumor effects in vitro, it was chosen for the in vivo experiments. LINC00662-depleted U87 cells were injected into the BALB/C immunodeficient mice. We found that the tumor volumes of the LINC00662 knockdown group were remarkably smaller than that of the H1 group after 21 days (Figure 2E). Meanwhile, the tumor weights displayed a similar result (Figure 2F and G). To further validate the inhibitory effects, IHC assay was used to assess the proliferation in the xenografts. Consequently, Ki-67 was significantly reduced in the LINC00662 knockdown group compared with the NC group (Figure 2H). Collectively, silenced LINC00662 suppressed glioma proliferation both in vitro and in vivo.
Silencing of LINC00662 Inhibited Cell Migration and EMT Pathway of Glioma
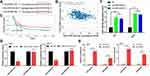
Since the previous study reported that LINC00662 regulated CRC metastasis,13 we wondered if LINC00662 displayed a similar function in glioma. Transwell assay showed that the decreased cell migration capacity following suppression of LINC00662 expression (Figure 3A). Similarly, the wound healing rate of LINC00662-silenced cells was significantly slower compared with that of control H1-treated glioma cells (Figure 3B). EMT is an important process to increase tumor cell migration capability.22 In support of EMT, two mesenchymal markers, N-cadherin and Vimentin were detected by Western blot. As a result, both were downregulated after LINC00662 knockdown in glioma cells (Figure 3C). Taken together, silencing LINC00662 suppressed migration ability and EMT pathway in glioma.
LINC00662 Acted as a ceRNA on miR-34a-5p in Glioma
Numerous studies suggest that lncRNAs could act as ceRNA that interfere with the function of miRNAs.23 Online analysis ENCORI and DIANA were used to predict that miR-34a-5p was the potential target of LINC00662 (Figure 4A), which was reported in prostate cancer.11 According to Pearson’s correlation coefficient analysis, the expression level of miR-34a-5p was negatively associated with LINC00662 expression in glioma tissues (Figure 4B). Then, we further analyzed the CGGA database to explore its effect on survival. The result indicated that the expression of miR-34a-5p was positively correlated with overall survival, consistent with the ceRNA hypothesis (Figure 4C). Next, we performed the dual-luciferase reporter assay to further validate the binding site. The results revealed that the transfection of miR-34a-5p mimics contributed to the significant decrease of luciferase activity of LINC00662-WT reporter, while the luciferase activity of LINC00662-MU reporter had no significant fluctuation in any group (Figure 4D). In addition, RIP assay indicated that LINC00662 was remarkably enriched by Ago2 antibody in U87 and U251 cells transfected with miR-34a-5p mimics in comparison with IgG group (Figure 4E). Moreover, qRT-PCR assay demonstrated that LINC00662 knockdown strikingly increased miR-34a-5p expression in U87 and U251 cells (Figure 4F). These data revealed that miR-34a-5p was a direct target of LINC00662 in glioma.
miR-34a-5p Exerted Anti-Tumor Effects on the Growth and Migration of Glioma Cells in vitro
Despite several studies have explored that miR-34a-5p could suppress tumorigenesis and progression of glioma,24–26 it is necessary to confirm the biological function of miR-34a-5p on the malignancy of glioma in our study. U87 and U251 cells were transfected with either miR-34a-5p mimics or miR-NC. Then, a series of gain-of-function experiments were conducted. Celltiter-Glo assay and flow cytometric analysis demonstrated that the ectopic miR-34a-5p expression markedly inhibited cell growth and induced apoptosis of U87 and U251 cells (Figure S2A and B). In addition, Transwell migration assay was performed to evaluate the motility of glioma cells after either miR-34a-5p or miR-NC transfection. The number of migratory miR-34a-5p-overexpressing U87 and U251 cells was much lower than those of the cells transfected with miR-NC (Figure S2C). Thus, miR-34a-5p functioned as an oncogene during glioma progression, in agreement with the previous studies.24,25
LMAN2L, a Direct Target of miR-34a-5p, Was Crucial for the Function of LINC00662
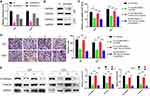
To investigate the target of miR-34a-5p regulated by LINC00662, we conducted bioinformatics analysis using four different algorithms, including ENCORI, TargetScan, miRDB and mirDIP (Table S1). From the Venn results, 12 potential miR-34a-5p target genes were identified (ARID4B, ATMIN, DAAM1, DCAF7, ELL2, FOXP1, GLCE, LMAN2L, METAP1, TMEM109, VAMP2, VCL) (Figure 5A). Among them, the expression levels of ARID4B, ATMIN, DCAF7, LMAN2L and METAP1 were negatively correlated with miR-34a-5p expression (Figures 5B and S3A). Furthermore, DCAF7 and LMAN2L were found to be upregulated in glioma tissues (Figures 5C and S3B). Meanwhile, DAAM1, VCL, TMEM109, GLCE and LMAN2L expression levels were correlated conversely with overall survival (Figures 5D and S3C). Then, expression analysis yielded that only LMAN2L was downregulated in U87 cells upon miR-34a-5p overexpression (Figure 5E). According to the bioinformatic analysis and qRT-PCR result, we supposed that LMAN2L is a direct target of miR-34a-5p. Next, we conducted the dual-luciferase assay to verify the predicted interaction between LMAN2L and miR-34a-5p (Figure 5F). Upregulation of miR-34a-5p markedly decreased the luciferase activity of plasmid LMAN2L-WT, which contains the wild-type miR-34a-5p binding site; however, the luciferase activity of the reporter plasmid containing the mutant binding site (LMAN2L-MU) showed no obvious change in U87 and U251 cells (Figure 5G). In addition, the expression of LMAN2L was downregulated with shLINC00662-1 alone and reversed by co-transfection with shLINC00662-1 and miR-34a-5p inhibitor (Figure 5H). Therefore, LMAN2L is a direct target of miR-34a-5p in glioma cells.
To verify the ceRNA network between LINC00662, miR-34a-5p and LMAN2L in glioma, we firstly manipulated LMAN2L expression using specifically targeting shRNAs. The knockdown efficiencies were evaluated by qRT-PCR and Western blot (Figure 6A and B). Next, Celltiter-Glo assay demonstrated that miR-34a-5p inhibitor could increase the growth of shLINC00662 glioma cells, which was reversed by LMAN2L knockdown (Figure 6C). Similarly, the anti-migratory effect and mesenchymal-epithelial transition (MET) induced by LINC00662 knockdown were rescued by miR-34a-5p inhibitor, while suppression of LMAN2L could reverse these effects (Figure 6D and E). Collectively, these data disclosed that LMAN2L and miR-34a-5p involved in LINC00662-mediated cell proliferation, migration and MET pathway.
Discussion
With the high occurrence and aggressive behavior, glioma has become a major reason for intracranial tumor relevant death.1 It is urgent and important to investigate the biological mechanism of glioma and find new therapeutic targets to prevent tumor cell growth and migration. Convincing evidence indicated that lncRNAs have been established as crucial regulators of glioma.27 Especially, LINC00662 is considered as an oncogene in several other tumors.28 We wondered whether LINC00662 played a similar role in glioma progression. In our study, we analyzed TCGA and CGGA databases to revealed that high LINC00662 expression level was correlated with unfavorable survival outcomes of glioma patients. By loss-of-function experiments, we demonstrated that suppression of LINC00662 inhibited the proliferation, mobility capacity and EMT pathway of glioma cells in vitro. The xenograft mouse model was a common method to evaluate the gene function in vivo. Consequently, xenograft and immunohistochemistry were adopted to verify the tumor-promoting role of LINC00662 in vivo.
It is well accepted that lncRNAs exert their regulatory functions by sequestration of miRNAs.29 Based on online databases, miR-34a-5p was predicted as a potential target of LINC00662 in glioma. Interestingly, miR-34a-5p was elucidated to be a direct target of LINC00662 in prostate cancer,11 as well as acting as a tumor suppressor in numerous cancers.30 We supposed LINC00662 and miR-34a-5p exerted a similar interaction in glioma. Next, correlation analysis between miR-34a-5p and LINC00662, together with the survival benefit of miR-34a-5p upregulation, gave us the confidence to further explore the ceRNA hypothesis. Then, the binding between LINC00662 and miR-34a-5p in glioma was verified using dual-luciferase reporter and RIP assay. Subsequently, functional experiments indicated that LINC00662 sponged miR-34-5p to promote glioma malignant phenotype, in agreement with the previous study.24
To investigate the target gene of the LINC00662/miR-34a-5p axis in glioma, we combined four bioinformatics algorithms (Targetscan, ENCORI, miRDB and mirDIP), TCGA and CGGA databases and found that LMAN2L was the downstream effector of the LINC00662/miR-34a-5p pathway. The lectins contain a diverse class of proteins, which are able to bind carbohydrates with considerable specificity. The selectin, a family member of lectin, has been confirmed to control lymphocyte homing and leukocyte trafficking to sites of inflammation.31,32 The galectins regulate cell–substratum interactions,33 contribute to the tumor-promoting microenvironment and lead to tumor cell metastasis.34 In addition, Cheng et al indicated LINC00662 regulated Claudin 8 (CLDN8) and Interleukin 22 (IL-22) co-expression to promote colon cancer metastasis,35 inferring that both LINC00662 and lectin family participated in neuroinflammation and tumor-promoting microenvironment.
LMAN2L, a transmembrane protein located at the endoplasmic reticulum, plays a crucial role in quality control of glycoproteins.36 LMAN2L gene mutation results in neurodevelopmental disorder, including intellectual disability and bipolar disorder.37,38 It is well documented that glycoproteins contribute to epithelial cell growth and malignant transformation.39,40 Therefore, we speculated that LMAN2L might contribute to glioma progression. However, to our best knowledge, there is no published study to investigate the relationship between LMAN2L and tumor malignancy, motivating us to explore the biological function of LMAN2L in glioma. As shown in Figure 7, LMAN2L was a direct target of miR-34a-5p regulated by LINC00662 evidenced by bioinformatic analysis and dual-luciferase reporter assay. Moreover, rescue experiments suggested that the interaction between the LINC00662/miR-34a-5p axis and LMAN2L is implicated in the progression of glioma and EMT pathway.
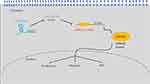 |
Figure 7 Schematic of the LINC00662/miR-34a-5p/LMAN2L/EMT regulatory network in glioma cells. |
Conclusion
In summary, our study indicated that suppression of LINC00662 attenuated cell proliferation and migration through activating miR-34a-5p to regulate the expression of LMAN2L and inhibiting EMT signaling pathway. These results may provide a novel perspective on the targeted therapy of glioma.
Funding
This study was supported by the Beijing Medical Research “Multi-center clinical big data study and multi-path tumorigenesis mechanisms and precision treatment research on brainstem glioma” (Grant No. 2018-7) and the National Natural Science Foundation of China (Grant No. 81872048).
Disclosure
Yibo Geng, Tian Li, and Liwei Zhang report grants from Beijing Medical Research and National Natural Science Foundation of China, during the conduct of the study. The authors report no other potential conflicts of interest in this work.
References
1. Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. doi:10.1093/neuonc/noz150
2. Wang T, Yang Y, Xu X, et al. An integrative survival analysis for multicentric low-grade glioma. World Neurosurg. 2020;134:e189–e195. doi:10.1016/j.wneu.2019.10.001
3. Li Y, Egranov SD, Yang L, Lin C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer. 2019;58(4):200–207. doi:10.1002/gcc.22691
4. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi:10.1534/genetics.112.146704
5. Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci. 2012;2(1):37. doi:10.1186/2045-3701-2-37
6. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
7. Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903.
8. Gong W, Su Y, Liu Y, Sun P, Wang X. Long non-coding RNA Linc00662 promotes cell invasion and contributes to cancer stem cell-like phenotypes in lung cancer cells. J Biochem. 2018;164(6):461–469. doi:10.1093/jb/mvy078
9. Liu Z, Yao Y, Huang S, et al. LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505(3):843–849. doi:10.1016/j.bbrc.2018.09.191
10. Xu D, Chen Y, Yuan C, Zhang S, Peng W. Long non-coding RNA LINC00662 promotes proliferation and migration in oral squamous cell carcinoma. Onco Targets Ther. 2019;12:647–656. doi:10.2147/OTT.S188691
11. Li N, Zhang LY, Qiao YH, Song RJ. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23(9):3688–3698.
12. Guo T, Gong C, Wu P, et al. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 2020;27(7):2191–2205. doi:10.1038/s41418-020-0494-3
13. Wang H, Yu M, Hu W, et al. Linc00662 promotes tumorigenesis and progression by regulating miR-497-5p/AVL9 axis in colorectal cancer. Front Genet. 2019;10:1385. doi:10.3389/fgene.2019.01385
14. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
15. Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Databaseissue):D92–97. doi:10.1093/nar/gkt1248
16. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi:10.7554/eLife.05005
17. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757
18. Tokar T, Pastrello C, Rossos AEM, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi:10.1093/nar/gkx1144
19. Xu C, Liu X, Geng Y, et al. Patient-derived DIPG cells preserve stem-like characteristics and generate orthotopic tumors. Oncotarget. 2017;8(44):76644–76655. doi:10.18632/oncotarget.19656
20. Kokovay E, Wang Y, Kusek G, et al. VCAM1 is essential to maintain the structure of the SVZ niche and acts as an environmental sensor to regulate SVZ lineage progression. Cell Stem Cell. 2012;11(2):220–230. doi:10.1016/j.stem.2012.06.016
21. Hu XL, Chen G, Zhang S, et al. Persistent expression of VCAM1 in radial glial cells is required for the embryonic origin of postnatal neural stem cells. Neuron. 2017;95(2):309–325. doi:10.1016/j.neuron.2017.06.047
22. Campbell K. Contribution of epithelial-mesenchymal transitions to organogenesis and cancer metastasis. Curr Opin Cell Biol. 2018;55:30–35. doi:10.1016/j.ceb.2018.06.008
23. Xu W, Yu S, Xiong J, Long J, Zheng Y, Sang X. CeRNA regulatory network-based analysis to study the roles of noncoding RNAs in the pathogenesis of intrahepatic cholangiocellular carcinoma. Aging. 2020;12(2):1047–1086. doi:10.18632/aging.102634
24. Ma S, Fu T, Zhao S, Gao M. MicroRNA-34a-5p suppresses tumorigenesis and progression of glioma and potentiates Temozolomide-induced cytotoxicity for glioma cells by targeting HMGA2. Eur J Pharmacol. 2019;852:42–50. doi:10.1016/j.ejphar.2019.03.005
25. Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H. NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci. 2018;11:225. doi:10.3389/fnmol.2018.00225
26. Di Bari M, Bevilacqua V, De Jaco A, et al. Mir-34a-5p mediates cross-talk between M2 muscarinic receptors and notch-1/EGFR pathways in U87MG glioblastoma cells: implication in cell proliferation. Int J Mol Sci. 2018;19(6):1631. doi:10.3390/ijms19061631
27. Huang Z, Zhao X, Wu X, et al. LncRNA UCA1 facilitated cell growth and invasion through the miR-206/CLOCK axis in glioma. Cancer Cell Int. 2019;19:316. doi:10.1186/s12935-019-1023-7
28. Wang CB, Wang Y, Wang JJ, Guo XL. LINC00662 triggers malignant progression of chordoma by the activation of RNF144B via targeting miR-16-5p. Eur Rev Med Pharmacol Sci. 2020;24(3):1007–1022.
29. Jiang J, Bi Y, Liu XP, et al. To construct a ceRNA regulatory network as prognostic biomarkers for bladder cancer. J Cell Mol Med. 2020;24:5375–5386. doi:10.1111/jcmm.15193
30. Wang X, Zhao Y, Lu Q, et al. MiR-34a-5p inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in esophageal squamous cell carcinoma by targeting LEF1 and inactivation of the hippo-YAP1/TAZ signaling pathway. J Cancer. 2020;11(10):3072–3081. doi:10.7150/jca.39861
31. Sinclair LV, Finlay D, Feijoo C, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9(5):513–521. doi:10.1038/ni.1603
32. Lee M, Kiefel H, LaJevic MD, et al. Transcriptional programs of lymphoid tissue capillary and high endothelium reveal control mechanisms for lymphocyte homing. Nat Immunol. 2014;15(10):982–995. doi:10.1038/ni.2983
33. Fan Y, Sun L, Yang S, He C, Tai G, Zhou Y. The roles and mechanisms of homogalacturonan and rhamnogalacturonan I pectins on the inhibition of cell migration. Int J Biol Macromol. 2018;106:207–217. doi:10.1016/j.ijbiomac.2017.08.004
34. Compagno D, Tiraboschi C, Garcia JD, et al. Galectins as checkpoints of the immune system in cancers, their clinical relevance, and implication in clinical trials. Biomolecules. 2020;10(5):750. doi:10.3390/biom10050750
35. Cheng B, Rong A, Zhou Q, Li LW. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J Exp Clin Cancer Res. 2020;39(1):5. doi:10.1186/s13046-019-1510-7
36. Qin SY, Kawasaki N, Hu D, Tozawa H, Matsumoto N, Yamamoto K. Subcellular localization of ERGIC-53 under endoplasmic reticulum stress condition. Glycobiology. 2012;22(12):1709–1720. doi:10.1093/glycob/cws114
37. Alkhater RA, Wang P, Ruggieri A, et al. Dominant LMAN2L mutation causes intellectual disability with remitting epilepsy. Ann Clin Transl Neurol. 2019;6(4):807–811. doi:10.1002/acn3.727
38. Chen DT, Jiang X, Akula N, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18(2):195–205. doi:10.1038/mp.2011.157
39. Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473(1):21–34. doi:10.1016/S0304-4165(99)00167-1
40. Chiu WC, Chiou TJ, Chung MJ, Chiang AN. β2-Glycoprotein I inhibits vascular endothelial growth factor-induced angiogenesis by suppressing the phosphorylation of extracellular signal-regulated kinase 1/2, Akt, and endothelial nitric oxide synthase. PLoS One. 2016;11(8):e0161950. doi:10.1371/journal.pone.0161950
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.