Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA LINC00511 Accelerates Proliferation and Invasion in Cervical Cancer Through Targeting miR-324-5p/DRAM1 Axis
Authors Zhang X, Wang Y, Zhao A, Kong F, Jiang L, Wang J
Received 23 March 2020
Accepted for publication 29 August 2020
Published 12 October 2020 Volume 2020:13 Pages 10245—10256
DOI https://doi.org/10.2147/OTT.S255067
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Xin Zhang,1 Yuyan Wang,2 Anqi Zhao,3 Fanshuang Kong,4 Lipeng Jiang,2 Jinfeng Wang5
1Department of Oncology, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China; 2Department of Obstetrics and Gynecology, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China; 3Capital Medical University, Beijing 100069, People’s Republic of China; 4Department of General Surgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China; 5Department of Pediatrics, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China
Correspondence: Jinfeng Wang
Department of Pediatrics, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou 121001, People’s Republic of China
Tel +86-13897836848
Email [email protected]
Purpose: Cervical cancer is the second most prevalent female malignance, and human papillomavirus (HPV) infection is the main pathogenic factor of cervical cancer. Emerging evidence has revealed that a number of long non-coding RNAs (lncRNAs) play critical roles in the tumorigenesis and progression of cervical cancer. The aim of this study was to further investigate the precise role of lncRNA LINC00511 in HPV-negative and HPV-positive cervical cancer cells and explore the potential regulatory mechanism.
Methods: The expression of LINC00511 in cervical cancer and cell lines was examined by RT-PCR. Fluorescence in situ hybridization analysis (FISH) assay was performed to detect the localization of LINC00511 in cervical cancer cells. Loss-of-function experiments of LINC00511 by siRNA interference were performed to assess its effects on HPV-negative and HPV-positive cervical cancer cells. Dual-luciferase reporter and RNA immunoprecipitation (RIP) assays were used to identify the target of LINC00511. Relative expression of related proteins was detected using Western blot.
Results: Herein, the results showed that LINC00511 was significantly up-regulated in cervical cancer and cell lines and mainly distributed in the cytoplasm of cervical cancer cells. Loss-of-function experiments indicated that silencing of LINC00511 inhibited the proliferation and invasion of both HPV-negative and HPV-positive cervical cancer cells, as well as promoted apoptosis by regulating the Bcl-2/Bax axis and Caspase 3 activation. Bioinformatic analysis, dual-luciferase reporter, and RIP assays showed that LINC00511 was a target of miR-324-5p, while DRAM1 was a direct target of miR-324-5p. The expression of miR-324-5p was down-regulated in cervical cancer, while the expression of DRAM1 was up-regulated. Moreover, the expression of LINC00511 was negatively correlated with miR-324-5p expression in cervical cancer tissues and positively correlated with DRAM1. Further, DRAM1 overexpression promoted both HPV-negative and HPV-positive cervical cancer cell proliferation and invasion, which could be reversed by miR-324-5p mimics or si-LINC00511.
Conclusion: Collectively, these results suggest that LINC00511 functions as a competing endogenous RNA (ceRNA) to regulate the miR-324-5p/DRAM1 axis, leading to HPV-negative and HPV-positive cervical cancer aggravation.
Keywords: cervical cancer, LINC00511, HPV-negative, HPV-positive, miR-324-5p, DRAM1
Introduction
Cervical cancer is the second most prevalent female malignance, with the highest incidence rate of gynecological cancer in recent years.1 In 2018, this malignance has caused 569,847 new cases and 311,365 deaths in the world.2 Although surgery and radiotherapy can effectively eliminate cervical tumors in situ, but for those in advanced stages, especially for patients with metastatic tumors, the effect is very limited.3 It is reported that the 5-year survival rate of patients with advanced cervical cancer is less than 40%, mainly due to tumor metastasis and recurrence.4,5 Human papillomavirus (HPV) infection is the main pathogenic factor of cervical cancer and the main content of early diagnosis, but nearly 15% of patients are HPV-negative.6,7 Therefore, further study is required to reveal the molecular mechanism of the occurrence and development of cervical cancer and identify novel therapeutic targets of HPV-positive and HPV-negative cervical cancer.
It has been substantially certified that long non-coding RNAs (lncRNAs), which are pervasively observed in the eukaryote genome, are frequently dysregulated in cancers.8 Plenty of lncRNAs regulate gene expression involving cancer cellular process, including proliferation, apoptosis, invasion, survival, and drug resistance, thereby participating in tumorigenesis and development.9,10 Previous studies have revealed a number of lncRNAs affect the growth and metastasis of cervical cancer, which means they can then serve as potential biomarkers or prognostic markers.11–14 LncRNA LINC00511 is reported to be up-regulated in various cancers, such as hepatocellular carcinoma,15 gastric cancer16 and ovarian cancer,17 exerting an oncogenic role in tumor progression.15–19 Recently, LINC00511 was revealed to be up-regulated in cervical cancer tissues and positively correlated with malignancy of cervical cancer, as well as poor prognosis of patients.20 In non-small cell lung cancer (NSCLC), LINC00511 has been reported to down-regulate LATS2 and KLF2 by binding to EZH1 and LSD1, promoting the progression of NSCLC.21 Jiang L et al reveal that silencing of LINC00511 could suppress TGF-β1-induced migration and invasion in lung cancer.22 Shi et al report that LINC00511 knockdown inhibits the proliferation and promotes apoptosis and autophagy of HeLa cells.23 However, it is unclear whether LINC00511 performs similar functions in cervical cancer cells, particularly in HPV-negative cells, as it has not been studied.
In the present study, we aimed to investigate the precise role of LINC00511 in both HPV-negative and HPV-positive cervical cancer cells, and explore the potential regulatory mechanism. Our data elucidated that LINC00511 accelerated the proliferation and invasion of both HPV-negative/-positive cervical cancer cells, and served as a competing endogenous RNA (ceRNA) to regulate DRAM1 by sponging up miR-324-5p.
Materials and Methods
Gene Expression Profiling Interactive Analysis (GEPIA)
The GEPIA (http://gepia.cancer-pku.cn/index.html) database is an interactive web server for analyzing the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline.24 We analyzed the expression of LINC00511 in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC; n = 306) tissues and normal tissues (n = 13).
Human Tissue Samples
A total of 19 pairs of cervical cancer and paracancerous tissues were collected from The First Affiliated Hospital of Jinzhou Medical University Hospital from March 2017 to April 2019. All specimens were frozen in liquid nitrogen and stored at −80 °C. All patients did not receive radiotherapy and chemotherapy before operation, and signed informed consent. This study was allowed by the Institutional Review Committee (No. KYLL202013).
Cell Culture and Transfection
Human cervical cancer cell lines SiHa (HPV-positive), CaSki (HPV-positive) and C33A (HPV-negative) and normal cervical epithelial cells (HUCEC) were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China), HEK293T cells were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were routinely cultured at 37 °C in DMEM medium (Gibco, USA) containing 10% FBS. The siRNA targeting LINC00511 (si-LINC00511; 50 nM; Cat#siB150715110235; Ribobio, Guangzhou, China) was transfected into cervical cancer cells to knock down its expression, siRNA control (50 nM; Cat#siN0000001; Ribobio) was used as negative control (NC), and cells without any treatment were mock group. miR-324-5p mimics (Cat# miR10000761; 50 nM), miR-NC (50 nM), pcDNA3.1-LINC00511 (2.5 μg), and pcDNA3.1-DRAM1 (2.5 μg) were also obtained from Ribobio.
Cells were incubated in a 6-well plate at a density of 3×105 cells/well and transfection was performed using lipofectamine 2000 (Invitrogen, USA) when cells had a confluence of 60–80%. After stable transfection for 24 h, cells were collected for further experiments. The sequence of si-LINC00511 was as follows: si-LINC00511#1, 5ʹ-GCTCTGGAGTAACTATTCC-3ʹ; si-LINC00511#2, 5ʹ-GTGGAGACAGAACGTTTGG-3ʹ. The sequence of siRNA control was 5ʹ-AATTCTCCGAACGTGTCACGT-3ʹ.
Quantitative Real‑Time PCR Analysis
Total RNA was isolated from cervical cancer cells or tissues using Trizol and reverse transcripted into cDNA using the cDNA Reverse Transcription Kit (Takara, Japan). The qRT-PCR reaction was performed using SYBR® Green Master Mix (TaKaRa). The obtained data were analyzed using the 2−ΔΔCt method, the relative expression of the target gene was normalized to the NC group. GAPDH or U6 was used as the control for LINC00511, DRAM1 or miR-324-5p.
Fluorescence in situ Hybridization Analysis (FISH)
The FISH analysis was performed using the FISH kit (Ribobio) and LINC00511 FISH probe (Ribobio) according to the instructions. Cover glasses were placed on the 24-well plate (6×104 cells/well) with 85% cell confluence. After being fixed with 4% paraformaldehyde at room temperature for 10 min, the cells were permeabilized with 0.5% Triton X-100 in PBS. Subsequently, the cells were incubated in pre-hybridization buffer at 37 °C for 30 min. Discarded pre-hybridization buffer and cells were incubated in hybridization buffer containing probes at 37 °C overnight in the dark. After washing 3 times with hybrid buffer at 42 °C in the dark, cells were stained with 4ʹ,6-diamidino-2-phenylindole for 10 min at room temperature in the dark. Finally, the coverslip was sealed with an anti-fluorescent quencher, and the fluorescence was observed under a fluorescence microscope (Olympus, Japan).
Cell Viability Assay
A Cell Counting Kit-8 (CCK8; Abcam, USA) was performed to assess cell viability. Cells were transfected with siRNAs or vectors for 24 h and then seeded in a 96-well plate at a density of 1000 cells/well. Cell viability was detected at 0, 24, 48, and 72 h, respectively. Before detection, cells were incubated with 10 μL of CCK8 reagent at 37 °C for 1.5 h. The OD value at 450 nm was measured using a microplate reader.
Colony Formation Assay
Following transfection for 24 h, cells were plated in 60-mm plates at a density of 500 cells/plate and incubated at 37 °C for 10 days. After being fixed with 4% paraformaldehyde for 30 min, cells were stained with 0.1% crystal violet for 30 min. The number of colonies were then counted.
Transwell Invasion Assay
Following transfection of 24 h, cells were suspended in serum-free medium and 500 μL of cell suspension (3×104 cells) were added in the upper chamber of Transwell chambers pre-coated with Matrigel (BD Biosciences, USA). The lower chambers were filled with 500 μL of medium containing 20% FBS. Following incubation for 24 h at 37 °C, the invaded cells were fixed with 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet for 20 min. The number of invaded cells was counted and imaged under an inverted microscope (Magnification X100; Olympus).
Cell Apoptosis Assay
Cell apoptosis was analyzed using the flow cytometry with an Annexin V-FITC/PI Apoptosis Detection kit (Solarbio, Beijing, China). Cervical cancer cells with a transfection of 24 h were cultured in serum-free medium for another 24 h. 1× 105 cells were stained with Annexin V/FITC for 5 min in the dark and stained with PI. The percentage of apoptotic cells was detected by a flow cytometry (FACScan, Beckton Dickinson) and then analyzed by Flowjo software.
Western Blot Analysis
Total protein was extracted from cervical cells transfected for 48 h using RIPA (CWBIO, Beijing, China) and determined by a BCA kit (CWBIO). The protein from each sample was separated by 10% SDS-PAGE and transferred to PVDF membranes. After being blocked with 5% defatted milk for 1 h, the membranes were incubated with primary antibodies (Proteintech, USA) at 4°C overnight. The members were then incubated with HRP-conjugated antibody for 1 h for immunoreaction, and the blot was visualized with an ECL Kit (CWBIO). Images were taken and analyzed by QUANTITY ONE software.
Dual-Luciferase Reporter Assay
The wild-type (wt) fragment of LINC00511 or DREAM1 was cloned into the luciferase reporter vector pmirGLO. The mutant (mut) fragment of LINC00511 or DREAM1 binding sites was also cloned into the luciferase reporter vector pmirGLO. After co-transfection with pmirGLO-LINC00511-wt/-mut or pmirGLO-DRAM1-wt/-mut and miR-324-5p mimics or miR-NC into HK293T cells for 48 h, the luciferase activity of each group was measured by the dual-luciferase reporter assay (Promega, USA).
RNA Immunoprecipitation (RIP)
An RNA-binding protein immunoprecipitation kit (Millipore, MA, USA) was used to unravel the interaction of LINC00511 and miR-324-5p to Ago2 protein in cervical cancer cells. Briefly, CaSki cells were lysed in RIP lysis buffer, cell lysates were then diluted with RIP buffer and incubated with anti-Ago2 antibody or anti-IgG coated on magnetic beads at 4 °C overnight. After being treated with Proteinase K at 55 °C for 30 min, the relative level of LINC00511 or miR-324-5p in the immunoprecipitate was measured by qRT-PCR assay.
Statistical Analysis
Data were expressed as the mean ± SD from more than three independent experiments and statistically analyzed by GraphPad Prism 7.0 software (GraphPad, CA, USA). Student’s t-test or one-way ANOVA was used to analyze the comparisons between groups. The relationship between LINC00511 and miR-324-5p or DRAM1 was analyzed by Pearson correlation analysis. P<0.05 was considered to be indicative of statistical significance.
Results
LINC00511 is Up-Regulated in Cervical Cancer Tissues and Cell Lines
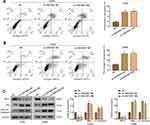
To preliminarily investigate the expression of LINC00511 in cervical cancer, we analyzed the expression of LINC00511 by the GEPIA.24 The results from the GEPIA database showed that the expression of LINC00511 in CESC tissues was significantly higher than that in normal tissues (Figure 1A). Subsequently, the expression of LINC00511 in 19 pairs of cervical cancer and paracancerous tissues was determined by qRT-PCR. As shown in Figure 1B, LINC00511 was significantly highly expressed in cervical cancer tissues compared with paracancerous tissues. In additionally, the expression of LINC00511 was remarkably up-regulated in HPV-positive SiHa and CaSki cells compared with HUCEC cells, and HPV-negative C33A cells also expressed a high level of LINC00511 (Figure 1C). Therefore, C33A and CaSki cells were selected for subsequent experiments due to their higher expression of LINC00511. Moreover, FISH assay showed that LINC00511 was mainly distributed in the cytoplasm of C33A and CaSki cells (Figure 1D).
Silencing of LINC00511 Suppresses the Proliferation and Invasion of Cervical Cancer Cells
To evaluate the roles of LINC00511 in cervical cancer, C33A and CaSki cells were transfected with siRNAs (si-LINC00511#1, si-LINC00511#2) to silence the expression of LINC00511. As detected by qRT-PCR, the expression of LINC00511 was significantly inhibited by si-LINC00511 in both C33A and CaSki cells (Figure 2A and B). Results of the CCK8 assay showed that silencing of LINC00511 significantly decreased the proliferation abilities of both C33A (Figure 2C) and CaSki cells (Figure 2D). A colony formation assay further confirmed that LINC00511 knockdown suppressed the colony formation ability of C33A (Figure 2E) and CaSki cells (Figure 2F). Moreover, the transwell assay revealed that silencing of LINC00511 significantly inhibited the invasion ability of C33A (Figure 2G) and CaSki (Figure 2H) cells compared with the corresponding control group. Collectively, these data suggested that LINC00511 exerted promoting effects on the proliferation and invasion of both HPV-positive and HPV-negative cervical cancer cells.
Silencing of LINC00511 Promotes Apoptosis in Cervical Cancer Cells
Flow cytometry assay illuminated that the percentage of apoptotic cells was significantly increased in both C33A (Figure 3A) and CaSki (Figure 3B) cells after LINC00511 was down-regulated. Further, results of Western blot analysis showed that the expression of anti-apoptotic protein Bcl-2 was down-regulated in C33A and CaSki cells following the silencing of LINC00511, while the expression of pro-apoptotic proteins Bax and cleaved Caspase 3 was up-regulated (Figure 3C). These data indicated that LINC00511 knockdown induced apoptosis in both HPV-positive and HPV-negative cervical cancer cells by regulating the Bcl-2/Bax axis and Caspase 3 activation. Additionally, due to the similar effects of si-LINC00511#1 and si-LINC00511#2, si-LINC00511#1 was used in the following experiments.
LINC00511 Serves as a Sponge of miR-324-5p in Cervical Cancer Cells
Since LINC00511 was mainly distributed in the cytoplasm of cervical cancer cells, inspired by the ceRNA hypothesis,25 we screened six candidate miRNAs (miR-150-5p, miR-105-5p, miR-324-5p, miR-624-3p, miR-2467-3p, and miR-4524-5p) which contained potential target sites for LINC00511 by Starbase. Dual-luciferase reporter assay showed that the luciferase activity of LINC00511 was significantly decreased in HEK293T cells transfected with miR-150-5p mimics, miR-324-5p mimics and miR-624-3p mimics (Figure 4A). Therefore, LINC00511 presented with the highest potential for binding to miR-324-5p. The binding sequence between LINC00511 (wild-type and mutant type) and miR-324-5p was shown in Figure 4B. Dual-luciferase reporter assays confirmed that miR-324-5p mimics obviously decreased the luciferase activity of LINC00511-wt, rather than LINC00511-mut (Figure 4C). RIP assay further manifested that LINC00511 and miR-324-5p were markedly enriched in Ago2-containing beads (Figure 4D). In addition, miR-324-5p expression was significantly increased in C33A and CaSki cells transfected with si-LINC00511 compared with the control group (Figure 4E). The expression of miR-324-5p was significantly down-regulated in cervical cancer tissues (Figure 4F). Further, Spearman correlation analysis showed that miR-324-5p expression was negatively correlated with LINC00511 in cervical cancer tissues (Figure 4G). Collectively, these findings suggest that LINC00511 serves as an endogenous sponge for miR-324-5p in cervical cancer cells.
DRAM1 is a Target Gene of miR-324-5p in Cervical Cancer Cells
By Starbase database, the potential targets for miR-324-5p were predicted as SP1, USP2, ETS1, DRAM1, and MUL1. By RT-PCR analysis, we found that the expression of ETS1 and DRAM1 mRNA was significantly decreased in CaSki cells transfected with miR-324-5p mimics, especially DRAM1 (Figure 5A). Therefore, DRAM1 was identified as a potential target gene of miR-324-5p, and the binding sites of them are shown in Figure 5B. Moreover, dual-luciferase reporter assay indicated that the luciferase activity of DRAM1-wt was markedly decreased by miR-324-5p mimics, rather than DRAM1-mut (Figure 5C). Western blot analysis showed that the expression of DRAM1 protein was down-regulated by miR-324-5p mimics and up-regulated by LINC00511 overexpression in both C33A and CaSki cells, which was restored in the presence of miR-324-5p mimics and pcDNA3.1-LINC00511 (Figure 5D and E). Compared with paracancerous tissues, the expression of DRAM1 mRNA was significantly up-regulated in cervical cancer tissues (Figure 5F). Further, the expression of DRAM1 was negatively correlated with miR-324-5p in cervical cancer tissues (Figure 5G), and positively correlated with LINC00511 (Figure 5H). Collectively, DRAM1 was a direct target gene of miR-324-5p in cervical cancer.
LINC00511 Promotes the Proliferation and Invasion of Cervical Cancer Cells by Sponging miR-324-5p and Regulating DRAM1 Expression
Rescue assays were carried out to further study the effects of the LINC00511/miR-324-5p/DRAM1 axis on cervical cancer cell's proliferation and invasion. CCK8 assay revealed that transfection with pcDNA3.1-DRAM1 significantly increased the viability of C33A and CaSki cells, while this promotion was significantly rescued by miR-324-5p mimics or LINC00511 knockdown (Figure 6A and B). Similarly, transwell assay showed that up-regulation of DRAM1 induced a significant increase of cell invasion in both C33A and CaSki cells, which was reversed by miR-324-5p mimics or si-LINC00511 (Figure 6C). Taken together, LINC00511 functions as a ceRNA to sponge miR-324-5p to regulate DRAM1 expression, promoting the proliferation and invasion of cervical cancer.
Discussion
The progression of cervical cancer is regulated by multiple factors, among which HPV persistent infection is the main factor.26 Therefore, it is necessary to further explore the pathogenesis of HPV-positive/negative cervical cancer to develop novel cervical cancer diagnosis and treatment methods. An increasing number of studies show that lncRNAs are involved in the occurrence and development of cervical cancer.13,27,28 As a newly identified lncRNA, LINC00511 has been reported to be up-regulated in a variety of cancers. Sun et al demonstrate that the abundance of LINC00511 is up-regulated in colorectal cancer tissues and cell lines, and LINC00511 overexpression promotes the growth of colorectal cancer cells.18 Wang D et al claim that LINC00511 expression is positively correlated with advanced clinical characters of gastric cancer, and inhibition of LINC00511 decreases tumor cell growth and invasion.16 Previous studies have revealed that LINC00511 is also up-regulated in cervical cancer and associated with poor prognosis, and serves as an oncogene gene in HPV-positive cervical cancer cells.20,23 Herein, what we have in common with previous research was that LINC00511 was significantly up-regulated in cervical cancer and promoted tumor development. In addition, this study revealed, for the first time, the promoting effect of LINC00511 on HPV-negative cervical cancer cells. Our data showed that the silencing of LINC00511 inhibited the proliferation and invasion of both HPV-positive and HPV-negative cells, promoted apoptosis by regulating the Bcl-2/Bax axis and Caspase 3 activation, indicating that LINC00511 acts as a tumor promoter in the progression of cervical cancer.
Shi et al reveal that LINC00511 regulates the expression of PLD1 via accumulation of transcription factor RXRA in the promoter region of the PLD1 in HeLa cells.23 In the present study, we confirmed that LINC00511 was mostly expressed in the cytoplasm of cervical cancer cells, which may function as a ceRNA. LINC00511 is also reported to amass in cytoplasm in gastric cancer29 and pancreatic adenocarcinoma cells.30 With the help of Starbase, dual-luciferase reporter, and RIP assays, our data disclosed that LINC00511 functioned as a sponge for miR-324-5p. Additionally, qRT-PCR analysis revealed that miR-324-5p expression was down-regulated in cervical cancer and negatively correlated with LINC00511 in cervical cancer. It has been reported that miR-324-5p inhibits the proliferation and invasion of colorectal cancer cells,31 and potentiates non-small cell lung cancer cell resistance to cisplatin.32 Jiang H et al report that miR-324-5p is down-regulated in cervical cancer tissues, inhibits cell proliferation and invasion, and acts as a downstream target of lncRNA TPT1-AS1.33 Therefore, miR-324-5p might act as a tumor suppressor in cervical cancer and be involved in the promoting effects of LINC00511.
To further explore the mechanism of LINC00511 in cervical cancer, further study was performed to find out the downstream target of LINC00511/miR-324-5p. Previous studies have reported the LINC00511/miR-625-5p/NFIX axis in gastric cancer,29 LINC00511/miR-195/EYA1 axis in hepatocellular carcinoma15 and LINC00511/miR-185/STXBP4 axis in breast cancer.34 Herein, we identified that DRAM1 was a direct target of miR-324-5p. Moreover, DRAM1 was significantly up-regulated in cervical cancer, its expression was negatively correlated with miR-324-5p expression in cervical cancer and positively correlated with LINC00511 expression, suggesting that DRAM1 may be involved in cervical cancer. DRAM1 is an evolutionarily conserved protein, which functions as a target for p53-mediated autophagy and programmed cell death, playing critical roles in the tumor suppressive effects of p53.35–37 DRAM1 can regulate apoptosis by regulating autophagy38 and increasing protein levels and lysosomal localization of Bax.39 Emerging evidence reveals that DRAM1 is involved in the progression of cancers.40–42 It has been revealed that DRAM1 regulates cell migration and invasion in glioblastoma stem cells by regulating autophagy.43 Chen C et al report that the silencing of DRAM1 inhibits the migration and invasion of HepG2 cells by autophagy-EMT pathway.40 Meng C et al indicate that DRAM1 functions as a target of miR-26b and inhibits irradiation‑induced autophagy in MCF-7 cells.41 Consistently, our study revealed the oncogenic role of DRAM1 in both HPV-positive and HPV-negative cervical cancer cells, up-regulation of DRAM1 enhanced the proliferation and invasion pf C33A and CaSki cells. Moreover, miR-324-5p mimics or the silencing of LINC00511 could counteract the promoting effects of DRAM1 on cell proliferation and invasion. Therefore, these results strongly suggest that LINC00511 functions as a ceRNA to regulate the miR-324-5p/DRAM1 axis in cervical cancer cells. The precise mechanism of DRAM1 in the proliferation and invasion of cervical cancer cells should be further explored in the future.
Conclusion
To sum up, our findings manifested that LINC00511 functions as an oncogene in both HPV-positive and HPV-negative cervical cancer cells via sponging miR-324-5p and regulating DRAM1 expression. LINC00511 is expected to be a potential therapeutic target for HPV-positive and HPV-negative cervical cancer. However, the oncogenic role of LINC00511 should be further explored in clinical analysis and in vivo in the future.
Funding
The present study was supported by the Natural Science Foundation of Science and Technology Department of Liaoning Province (Grant No. 20180530098, 20170540338, and 20170540340), and the Natural Science Foundation of Education Department of Liaoning Province (Grant No. JYTQN201713).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
3. Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35.
4. Smith R, Cokkinides V, Brawley OW. Cancer screening in the United States, 2011. CA Cancer J Clin. 2011;59(1):27–41. doi:10.3322/caac.20008
5. Dou X, Zhou Q, Wen M, et al. FOXD2-AS1Long noncoding RNA promotes the malignancy of cervical cancer by sponging microRNA-760 and upregulating hepatoma-derived growth factor. Front Pharmacol. 2019;10:1700.
6. Chelimo C, Wouldes TA, Cameron LD, Elwood JM. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J Infect. 2013;66(3):207–217. doi:10.1016/j.jinf.2012.10.024
7. Wang HY, Kim G, Cho H, et al. Diagnostic performance of HPV E6/E7, hTERT, and Ki67 mRNA RT-qPCR assays on formalin-fixed paraffin-embedded cervical tissue specimens from women with cervical cancer. Exp Mol Pathol. 2015;98(3):510–516. doi:10.1016/j.yexmp.2015.03.036
8. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
9. Wu Y, Shao A, Wang L. The role of lncRNAs in the distant metastasis of breast cancer. Front Oncol. 2019;9:407. doi:10.3389/fonc.2019.00407
10. Mai H, Zhou B, Liu L. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):198. doi:10.1186/s13046-019-1213-0
11. Yang W, Xu X, Hong L, Wang Q, Huang J, Jiang L. Upregulation of lncRNA GAS5 inhibits the growth and metastasis of cervical cancer cells. J Cell Physiol. 2019;234(12):23571–23580. doi:10.1002/jcp.28926
12. Zhang -J-J, Fan L-P. Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomed Pharmacother. 2019;117:108726. doi:10.1016/j.biopha.2019.108726
13. Wang Q, Yan S-P, Chu D-X. Silencing of long non-coding RNA RP1-93H18.6 acts as a tumor suppressor in cervical cancer through the blockade of the PI3K/Akt axis. Mol Ther Nucleic Acids. 2020;19:304–317. doi:10.1016/j.omtn.2019.10.041
14. Luo W, Wang M, Liu J, Cui X, Wang H. Identification of a six lncRNAs signature as novel diagnostic biomarkers for cervical cancer. J Cell Physiol. 2020;235(2):993–1000. doi:10.1002/jcp.29015
15. Hu W-Y, Wei H-Y, Li K-M, Wang R-B, Xu X-Q, Feng R. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed Pharmacother. 2020;121:109642. doi:10.1016/j.biopha.2019.109642
16. Wang D, Liu K, Chen E. LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol Biol Lett. 2020;25(1):4. doi:10.1186/s11658-020-0201-x
17. Wang K, Zhu G, Bao S, Chen S. Long non-coding RNA LINC00511 mediates the effects of ESR1 on proliferation and invasion of ovarian cancer through miR-424-5p and miR-370-5p. Cancer Manag Res. 2019;11:10807–10819. doi:10.2147/CMAR.S232140
18. Sun S, Xia C, Xu Y. HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed Pharmacother. 2020;125:110014. doi:10.1016/j.biopha.2020.110014
19. Du X, Tu Y, Liu S. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J Cell Mol Med. 2020;24(2):1474–1487. doi:10.1111/jcmm.14829
20. Yu CL, Xu XL, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep. 2019;39(9).
21. Zhu FY, Zhang SR, Wang LH, Wu WD, Zhao H. LINC00511 promotes the progression of non-small cell lung cancer through downregulating LATS2 and KLF2 by binding to EZH2 and LSD1. Eur Rev Med Pharmacol Sci. 2019;23(19):8377–8390.
22. Jiang L, Xie X, Bi R, Ding F, Mei J. Knockdown of Linc00511 inhibits TGF-β-induced cell migration and invasion by suppressing epithelial-mesenchymal transition and down-regulating MMPs expression. Biomed Pharmacother. 2020;125:109049. doi:10.1016/j.biopha.2019.109049
23. Shi Y, Liu M, Huang Y, Zhang J, Yin L. Promotion of cell autophagy and apoptosis in cervical cancer by inhibition of long noncoding RNA LINC00511 via transcription factor RXRA-regulated PLD1. J Cell Physiol. 2020.
24. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
25. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
26. Schiffman M, Castle P, Jeronimo J, Rodriguez AC, Wacholder S. Human papilloma virus and cervical cancer. Lancet. 2007;370(9590):890–907. doi:10.1016/S0140-6736(07)61416-0
27. Liu Y, Yang Y, Li L. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol. 2018;96(1):38–43. doi:10.1139/bcb-2017-0188
28. Feng S, Liu W, Bai X. LncRNA-CTS promotes metastasis and epithelial-to-mesenchymal transition through regulating miR-505/ZEB2 axis in cervical cancer. Cancer Lett. 2019;465:105–117. doi:10.1016/j.canlet.2019.09.002
29. Chen Z, Wu H, Zhang Z, Li G, Liu B. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. 2019;19(1):351. doi:10.1186/s12935-019-1070-0
30. Zhao X, Liu Y, Li Z. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018;22(1):655–667. doi:10.1111/jcmm.13351
31. Gu C, Zhang M, Sun W, Dong C. Upregulation of miR-324-5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol Res. 2019;27(5):515–524. doi:10.3727/096504018X15166183598572
32. Ba Z, Zhou Y, Yang Z, Xu J, Zhang X. miR-324-5p upregulation potentiates resistance to cisplatin by targeting FBXO11 signalling in non-small cell lung cancer cells. J Biochem. 2019;166(6):517–527. doi:10.1093/jb/mvz066
33. Jiang H, Huang G, Zhao N. Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. J Exp Clin Cancer Res. 2018;37(1):169. doi:10.1186/s13046-018-0846-8
34. Liu L, Zhu Y, Liu AM, Feng Y, Chen Y. Long noncoding RNA LINC00511 involves in breast cancer recurrence and radioresistance by regulating STXBP4 expression via miR-185. Eur Rev Med Pharmacol Sci. 2019;23(17):7457–7468.
35. Crighton D, Wilkinson S, O’Prey J. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi:10.1016/j.cell.2006.05.034
36. Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3(1):72–74. doi:10.4161/auto.3438
37. Criollo A, Dessen P, Kroemer G. DRAM: a phylogenetically ancient regulator of autophagy. Cell Cycle. 2009;8(15):2319–2320. doi:10.4161/cc.8.15.9153
38. Zhang X. Mechanism of DRAM1’s regulation of autophagy and apoptosis. J Pharmacogenomics Pharmacoproteomics. 2015;06.
39. Guan -J-J, Zhang X-D, Sun W, Qi L, Wu J-C, Qin Z-H. DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX. Cell Death Dis. 2015;6(1):e1624. doi:10.1038/cddis.2014.546
40. Chen C, Liang QY, Chen HK, et al. DRAM1 regulates the migration and invasion of hepatoblastoma cells via autophagy-EMT pathway. Oncol Lett. 2018;16(2):2427–2433.
41. Meng C, Liu Y, Shen Y, et al. MicroRNA-26b suppresses autophagy in breast cancer cells by targeting DRAM1 mRNA, and is downregulated by irradiation. Oncol Lett. 2018;15(2):1435–1440.
42. Lu T, Zhu Z, Wu J, et al. DRAM1 regulates autophagy and cell proliferation via inhibition of the phosphoinositide 3-kinase-Akt-mTOR-ribosomal protein S6 pathway. Cell Commun Signal. 2019;17(1):28. doi:10.1186/s12964-019-0341-7
43. Galavotti S, Bartesaghi S, Faccenda D. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32(6):699–712. doi:10.1038/onc.2012.111
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.