Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA HULC Promotes Cervical Cancer Cell Proliferation, Migration and Invasion via miR-218/TPD52 Axis
Authors Lu W, Wan X, Tao L, Wan J
Received 29 September 2019
Accepted for publication 12 December 2019
Published 5 February 2020 Volume 2020:13 Pages 1109—1118
DOI https://doi.org/10.2147/OTT.S232914
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Wenjun Lu,1 Xiaobin Wan,2 Limin Tao,1 Junhui Wan1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Nanchang University, Nanchang 330006, Jiangxi, People’s Republic of China; 2Department of General Surgery, The Third Hospital of Nanchang University, Nanchang 330006, Jiangxi, People’s Republic of China
Correspondence: Limin Tao; Junhui Wan
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Nanchang University, No. 17 Yongwai Zhengjie, Donghu District, Nanchang 330006, Jiangxi, People’s Republic of China
Tel +86-13879136013; +86-13970974546
Email [email protected]; [email protected]
Objective: Long non-coding RNAs (lncRNAs) have been identified as important players in tumorigenesis. LncRNA highly upregulated in liver cancer (HULC) has been identified as a key regulator in the progression of various cancers. However, the functional role and the mechanisms of HULC in regulating cervical cancer cell behavior remain unclear.
Methods: HULC expression, miR-218 expression and TPD52 mRNA level in cervical cancer cells were examined by qRT-PCR. Cell proliferation was evaluated by MTT assay. Cell migration and invasion were examined by Transwell assay. TPD52 protein level was measured by Western blot. Dual-luciferase reporter assay was measured to verify the combination of HULC and miR-218 as well as miR-218 and TPD52.
Results: HULC expression was upregulated in cervical cancer cell lines, and HULC promoted cervical cancer cell proliferation, migration and invasion. Mechanistically, HULC acted as a sponge of miR-218 to elevate expression of TPD52, a target of miR-218, and thereby promoted cervical cancer cell proliferation, migration, and invasion.
Conclusion: HULC promotes cervical cancer cell proliferation, migration and invasion via miR-218/TPD52 axis.
Keywords: HULC, miR-218, TPD52, cervical cancer cell proliferation
Introduction
Cervical cancer is one of the most common malignancies in the female reproductive system.1 However, the initial stages of cervical cancer are usually asymptomatic.2 Thus, a certain number of specific and sensitive non-invasive biomarkers are urgently needed to predict the prognosis of cervical cancer.3
Long non-coding RNAs (lncRNAs) are a group of RNAs greater than 200 nucleotides in length. Increasing evidence indicates that lncRNAs play important roles in regulating various cellular processes.4 It has been reported that there are several lncRNAs involved in cervical cancer development.5–7 LncRNAs play a role in the process of apoptosis of cervical cancer cells, tumor invasion and metastasis. So far, only a small fraction of lncRNA has been characterized in detail.8 Some lncRNAs regulate important cancer processes, including proliferation, migration, and invasion and drug resistance.9 More lncRNAs that affect cancer-related gene expression still need to be identified.
Highly upregulated in liver cancer (HULC) is a lncRNA that has recently been identified as a key regulator in the progression of various cancers.10 Wang et al revealed that the expression of HULC was upregulated in cervical cancer, and associated with overall survival,11 however, the effect and regulatory mechanism of HULC on proliferation, migration and invasion of cervical cancer cells remain unclear. MicroRNA-218 (miR-218) is a tumor-suppressive miRNA in cancers. MiR-218 was downregulated in cervical cancer, and miR-218 overexpression was found to inhibit cervical cancer cell viability, cell growth and metastasis, and promote apoptosis.12,13 Bioinformatics using miRanda predicted that HULC and miR-218 have partially complementary sequences, suggesting that HULC may function as a “miRNA sponge” of miR-218. The prediction of target genes using TargetScan showed that there were binding sites between miR-218 and 3ʹ-UTR of the oncogenic tumor protein D52 (TPD52) mRNA.14 Therefore, we speculated that HULC might competitively bind with miR-218 to regulate the TPD52 expression. In the current study, we aimed to examine the role and molecular mechanisms of HULC in regulating cervical cancer cell behavior.
Materials and Methods
Cell Lines and Cell Culture
Human cervical epithelial cells (H8 cells) and cervical cancer cells (HeLa, SiHa, CaSki and C-33A cells) purchased from Shanghai Institute of Cell Biology (Shanghai, China) were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin/streptomycin (Beijing Solarbio Science & Technology Co., China) with 5% CO2 at 37°C. The cells cultured to logarithm phase were used in the following experiments. The expression of HULC was detected in the above cell lines. The HULC overexpression vector, HULC siRNA (si-HULC), TPD52 overexpression vector, si-TPD52, miR-218 mimic, miR-218 inhibitor and their controls were synthesized by GenePharma (Shanghai, China) and then, respectively, transfected to cervical cancer cells using Lipofectamine 2000 (Invitrogen, USA). Finally, 48 hrs after transfection, transfected cells were collected and used in further experiments.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from cultured cells using Trizol reagent (Invitrogen, USA) and reverse-transcribed to cDNA using a PrimeScript RT Reagent Kit (TaKaRa, China) following the manufacturer’s protocol. qRT-PCR was performed to amplify the cDNA template using the SYBR Green PCR kit (TaKaRa, China). The levels of HULC and miR-218 were normalized to those of U6. The mRNA level of TPD52 was normalized to GAPDH. Specific PCR primers were synthesized at Invitrogen, USA. The relative expression was calculated using the 2−ΔΔCT method.
Western Blot
Total proteins were extracted from cervical cancer cell lines using RIPA lysis buffer (Beyotime, China). Equal amounts of protein were separated by 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes. After blocking with 5% skim milk, the membranes were incubated overnight at 4°C with anti-TPD52 antibody (1:500; Santa Cruz Biotechnology, Inc, USA), followed by incubation with horseradish peroxidase-conjugated secondary antibody (Boster, China) at room temperature for 2 hrs. The immunoreactive bands were detected using Electrochemiluminescence Detection Kit (Thermo Fisher Scientific, USA). β-actin (Boster, China) served as the loading control.
Cell Proliferation
Cells were seeded into 96-well plates at 2.5 × 103 cells per well. After seeding, the MTT assay was performed according to the manufacturer’s instructions. The optical density (OD) values at 490 nm were detected by an enzyme-labeled analyzer and were normalized to control well. All experiments were performed three times.
Cell Migration and Invasion Assay
For cell migration assay, serum-starved cells were suspended and plated into the upper chambers of 24-well Transwell plates with 8.0-μm pore. Full-serum medium containing DMEM with 10% FBS was then added into the lower chambers as the chemoattractant. After 24 hrs of incubation, cells were washed with PBS, fixed with 4% formaldehyde for 15 mins at room temperature and stained with 0.5% crystal violet for 20 mins. Finally, migratory cells were counted from 10 different fields of each filter. Cell invasion assay was performed the same way as the cell migration assay as described above, except that the upper chambers of 24-well Transwell plates were precoated with Matrigel (BD Biosciences, USA) membrane.
Luciferase Reporter Assay
The sequences of HULC and TPD52 3′-UTR were amplified from normal human genomic DNA and subcloned into the pmirGLO luciferase reporter vector (Promega, USA). HeLa cells were co-transfected with wild-type (WT) or mutant (Mut) 3′-UTR vectors and miR-218 mimic using Lipofectamine 2000. Then, the Dual-Luciferase Reporter Assay System (Promega) was used to conduct Luciferase reporter assay according to the manufacturer’s protocol. The luciferase activities were normalized to Renilla luciferase activity. All experiments were performed three times.
Statistical Analysis
All statistical analyses were performed using SPSS version 16.0 (SPSS, Inc., Chicago, USA). p<0.05 was considered to indicate a statistically significant difference. The unpaired Student’s t-test was used to analyze the differences between the two groups. One-way analysis of variance (ANOVA) was used to analyze differences among two or three groups.
Results
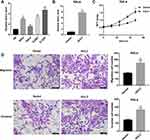
HULC Promotes Cervical Cancer Cell Proliferation, Migration and Invasion
QRT-PCR was used to detect the expression levels of HULC in cervical cancer cell lines such as HeLa, SiHa, CaSki and C-33A. The results demonstrated that HULC level was significantly upregulated in cervical cancer cells when compared with that in cervical epithelial H8 cells (Figure 1A). By performing the qRT-PCR assay, we found that after HULC overexpression plasmid has been transfected into cervical cancer HeLa cells, HULC expression was sharply upregulated in the overexpression group when compared with the vector group (Figure 1B). Subsequently, we analyzed the biological functions of HULC in HeLa cells. Forty-eight hours after transfection, cell proliferation was measured by using MTT assay. Results showed that HULC overexpression significantly promoted HeLa cell proliferation (Figure 1C). Meanwhile, Transwell assay was performed to investigate the role of HULC in regulating cell migration and invasion in HeLa cells. Data revealed that the number of migrating cells in the HULC overexpression group was significantly increased compared with the vector group (Figure 1D), and cell invasion showed a similar pattern in HeLa cells (Figure 1D). To further confirm the role of HULC in cervical cell proliferation, migration and invasion, HULC siRNA (si-HULC) was transfected into C-33A cells and the silencing efficiency of HULC was subsequently confirmed by qRT-PCR analysis, HULC expression was lower in the si-HULC group than that in the si-Ctrl group (Figure 2A). The results from the MTT proliferation assay showed that cell proliferation was significantly inhibited after si-HULC transfection in C-33A cells (Figure 2B). As shown in Figure 2C, compared with the si-Ctrl group, the decreased number of migrated and invaded cells was observed in the si-HULC group. These observations indicate that HULC overexpression promotes, whereas HULC silencing inhibits cervical cancer cell proliferation, migration and invasion.
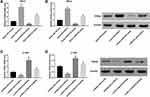
HULC Directly Targets miR-218, Which Targets TPD52
Next, we explored the molecular mechanism by which HULC promotes proliferation, migration and invasion in cervical cancer cells. We used miRanda (http://www.microrna.org/microrna/home.do) to predict the putative miRNA targets of lncRNA HULC and found that there was a highly conservative and specific combination sequence between HULC and miR-218 (Figure 3A). Our results showed that miR-218 mimic significantly repressed luciferase activity when co-transfected with a reporter containing WT HULC but not Mut HULC (Figure 3A). Interestingly, TargetScan analysis (http://www.targetscan.org/vert_72/) revealed that TPD52 was identified as a putative target of miR-218. The WT and Mut 3′-UTR of TPD52 were inserted into a luciferase reporter vector. Data showed that miR-218 could significantly inhibit luciferase expression of Luc-TPD52-WT. Mutation at any of the two sites could partly abrogate the suppressing effect (Figure 3B). After HULC overexpression plasmid was transfected into HeLa cells, qRT-PCR analysis exhibited that HULC overexpression significantly suppressed miR-218 expression (Figure 3C) but markedly promoted TPD52 mRNA and protein levels in HeLa cells (Figure 3D and E). In addition, upregulated miR-218 level (Figure 3F) and downregulated TPD52 mRNA and protein level (Figure 3G and H) were observed in C-33A cells transfected with si-HULC relative to those in C-33A cells transfected with siRNA control. These data indicate that HULC directly targets miR-218, which targets TPD52.
HULC Regulates TPD52 Expression by Regulating miR-218
To identify the role of HULC in regulating miR-218 and TPD52 expression, HeLa cells were co-transfected with HULC overexpression vector and miR-218 mimic. We examined the expression level of TPD52 in transfected cells by qRT-PCR and Western blot. The results showed that TPD52 expression both at mRNA level and protein level were significantly upregulated in HeLa cells transfected with HULC overexpression, but these effects were reversed by miR-218 mimic (Figure 4A and B). On the other hand, C-33A cells were co-transfected with si-HULC and miR-218 inhibitor. Data revealed that HULC silencing decreased TPD52 mRNA and protein levels, and these effects were reversed after miR-218 expression in C-33A cells was inhibited (Figure 4C and D). From these results, it is clear that HULC regulates TPD52 expression via sponging miR-218 in cervical cancer cells.
HULC Promotes Cervical Cancer Cell Proliferation, Migration and Invasion via miR-218/TPD52 Axis
Finally, we sought to elucidate whether miR-218/TPD52 axis was involved in the HULC-regulated cervical cancer cell proliferation, migration and invasion. To this end, HeLa cells were co-transfected with HULC overexpression vector and miR-218 mimic or si-TPD52. As shown in Figure 5A–C, the ability of HULC overexpression to promote proliferation, migration and invasion was markedly compromised when miR-218 expression in HeLa cells was upregulated. Furthermore, MTT and Transwell assay results indicated that HULC promoted the cell proliferation, migration and invasion, and these effects could be reversed by TPD52 silencing (Figure 5D–F). To further address this issue, C-33A cells were co-transfected with si-HULC and TPD52 overexpression vector. Data revealed that TPD52 overexpression effectively abrogated the HULC silencing-mediated inhibition of C-33A cell proliferation, migration, and invasion (Figure 6A–C). Taken together, these results suggest that HULC promotes cervical cancer cell proliferation, migration and invasion via miR-218/TPD52 axis.
Discussion
In the present work, we report that HULC expression was upregulated in cervical cancer cell lines, and HULC promoted cervical cancer cell proliferation, migration and invasion. Mechanistically, HULC acted as a sponge of miR-218 to elevate expression of TPD52, a target of miR-218, and thereby promoted cervical cancer cell proliferation, migration, and invasion.
As an oncogene, HULC has also been proved to play a critical role in regulating tumor metastasis in colorectal cancer,15 hepatocellular carcinoma,16 and bladder cancer.17 Wang et al revealed that the expression of HULC was upregulated in cervical cancer, and associated with overall survival,11 indicating that HULC may serve as a predictive biomarker for the prognosis of cervical cancer. In the present study, our data also demonstrated that HULC had high expression in cervical cancer cells, which is consistent with the findings showing HULC upregulation in cervical cancer tissues.11 Besides, our results showed that overexpression of HULC significantly promoted cervical cancer cell proliferation, migration and invasion. Our present study further supplements the oncogenic function of HULC in cervical cancer based on the existing literature and supports the view that HULC could act as a potential cervical cancer marker.
miR-218 has been shown to confer tumor-suppressive function in some cancers, such as liver cancer,18 oral cancer,19 bladder cancer,20 and nasopharyngeal cancer.21 With regard to cervical cancer, a previous study demonstrated that miR-218 was decreased in cervical cancer tissues, which was associated with tumor progression and poor prognosis.22 Subsequent research provided evidence that abundant miR-218 increased the radiosensitivity in cervical cancer cells.23 More recently, Zhu et al12,13 reported that miR-218 also produced anti-tumor effects on cervical cancer cells in vitro. In line with this, our results showed that miR-218 mimics significantly inhibited cervical cancer cell proliferation, migration and invasion. These results suggested that miR-218 plays a tumor-suppressive role in cervical cancer. TPD52 is a recently identified proto-oncogene, which was reported to be upregulated in several types of cancers.24,25 Wu et al reported that inhibition of TPD52 could suppress cervical cancer cell proliferation and induce cell apoptosis.26 Consistent with this, our results showed that TPD52 silencing inhibited, whereas TPD52 overexpression promoted cervical cell proliferation, migration, and invasion.
LncRNAs have been well known to exert their functions by acting as a ceRNA to segregate miRNAs away from target mRNAs, leading to the upregulation of miRNA target genes and suppression of miRNAs-mediated functional roles.27,28 Using bioinformatics analysis and luciferase reporter assay, we found that HULC acted as a sponge of miR-218 by binding to miR-218. Furthermore, TPD52 was confirmed as a direct target of miR-218, which was consistent with a previous study showing that miR-218 inhibited prostate cancer cell growth and promotes apoptosis by targeting and repressing TPD52 expression.14 Of note, we then found that the oncogenic function of HULC was abrogated when miR-218 expression was upregulated or TPD52 was downregulated in cervical cancer cells. Moreover, the tumor-suppressive effect of HULC silencing in cervical cancer cells could be attenuated by TPD52 overexpression. These data indicated that HULC promotes cervical cancer cell proliferation, migration and invasion via miR-218/TPD52 axis.
Conclusion
In summary, we demonstrated that HULC acts as a sponge of miR-218 to elevate expression of TPD52, a target of miR-218, and thereby promotes cervical cancer cell proliferation, migration, and invasion. Collectively, our findings provide a new perspective that the regulatory network of different lncRNAs may act as a critical role in cervical cancer progress.
Disclosure
Wenjun Lu and Xiaobin Wan are first coauthors. The authors declare no conflicts of interest.
References
1. Flanagan MB. Primary high-risk human papillomavirus testing for cervical cancer screening in the united states: is it time? Arch Pathol Lab Med. 2018;142(6):688–692. doi:10.5858/arpa.2018-0001-RA
2. Kim BW, Cho H, Choi CH, et al. Clinical significance of OCT4 and SOX2 protein expression in cervical cancer. BMC Cancer. 2015;15:1015. doi:10.1186/s12885-015-2015-1
3. Pedroza-Torres A, Fernandez-Retana J, Peralta-Zaragoza O, et al. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 2016;142(3):557–565. doi:10.1016/j.ygyno.2016.07.093
4. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi:10.1146/annurev-biochem-051410-092902
5. Ji F, Wuerkenbieke D, He Y, Ding Y, Du R. Long noncoding RNA HOTAIR: an oncogene in human cervical cancer interacting with MicroRNA-17-5p. Oncol Res. 2018;26(3):353–361. doi:10.3727/096504017X15002869385155
6. Yan Q, Tian Y, Hao F. Downregulation of lncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol Res. 2018. doi:10.3727/096504018X15185714083446
7. Yu X, Yang Y, Li Y, et al. Baicalein inhibits cervical cancer progression via downregulating long noncoding RNA BDLNR and its downstream PI3K/Akt pathway. Int J Biochem Cell Biol. 2018;94:107–118. doi:10.1016/j.biocel.2017.11.009
8. Li Y, Wang Z, Wang Y, et al. Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget. 2016;7(29):45027–45041. doi:10.18632/oncotarget.7801
9. Liao T, Qu N, Shi RL, et al. BRAF-activated LncRNA functions as a tumor suppressor in papillary thyroid cancer. Oncotarget. 2017;8(1):238–247. doi:10.18632/oncotarget.10825
10. Yu X, Zheng H, Chan MT, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017;21(2):410–417. doi:10.1111/jcmm.12956
11. Wang YF, Zhang S, Li XQ, Wang Y. Expression of lncRNA HULC in cervical cancer and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20(19):3987–3991.
12. Zhu L, Tu H, Liang Y, Tang D. MiR-218 produces anti-tumor effects on cervical cancer cells in vitro. World J Surg Oncol. 2018;16(1):204. doi:10.1186/s12957-018-1506-3
13. Xu Y, He Q, Lu Y, Tao F, Zhao L, Ou R. MicroRNA-218-5p inhibits cell growth and metastasis in cervical cancer via LYN/NF-κB signaling pathway. Cancer Cell Int. 2018;18. doi:10.1186/s12935-018-0673-1
14. Han G, Fan M, Zhang X. microRNA-218 inhibits prostate cancer cell growth and promotes apoptosis by repressing TPD52 expression. Biochem Biophys Res Commun. 2015;456(3):804–809. doi:10.1016/j.bbrc.2014.12.026
15. Shaker OG, Senousy MA, Elbaz EM. Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci Rep. 2017;7(1):16246. doi:10.1038/s41598-017-16500-4
16. Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi:10.1038/onc.2016.521
17. Wang J, Ma W, Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20(4):425–434. doi:10.3233/CBM-170188
18. Fu WM, Tang LP, Zhu X, et al. MiR-218-targeting-Bmi-1 mediates the suppressive effect of 1,6,7-trihydroxyxanthone on liver cancer cells. Apoptosis. 2015;20(1):75–82. doi:10.1007/s10495-014-1047-3
19. Uesugi A, Kozaki K, Tsuruta T, et al. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011;71(17):5765–5778. doi:10.1158/0008-5472.CAN-11-0368
20. Tatarano S, Chiyomaru T, Kawakami K, et al. miR-218 on the genomic loss region of chromosome 4p15.31 functions as a tumor suppressor in bladder cancer. Int J Oncol. 2011;39(1):13–21. doi:10.3892/ijo.2011.1012
21. Alajez NM, Lenarduzzi M, Ito E, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71(6):2381–2391. doi:10.1158/0008-5472.CAN-10-2754
22. Yu J, Wang Y, Dong R, Huang X, Ding S, Qiu H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol. 2012;138(4):671–674. doi:10.1007/s00432-012-1147-9
23. Yuan W, Xiaoyun H, Haifeng Q, et al. MicroRNA-218 enhances the radiosensitivity of human cervical cancer via promoting radiation induced apoptosis. Int J Med Sci. 2014;11(7):691–696. doi:10.7150/ijms.8880
24. Rubin MA, Varambally S, Beroukhim R, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64(11):3814–3822. doi:10.1158/0008-5472.CAN-03-3881
25. Byrne JA, Balleine RL, Schoenberg Fejzo M, et al. Tumor protein D52 (TPD52) is overexpressed and a gene amplification target in ovarian cancer. Int J Cancer. 2005;117(6):1049–1054. doi:10.1002/(ISSN)1097-0215
26. Wu Y, Huang J, Xu H, Gong Z. Over-expression of miR-15a-3p enhances the radiosensitivity of cervical cancer by targeting tumor protein D52. Biomed Pharmacother. 2018;105:1325–1334. doi:10.1016/j.biopha.2018.06.033
27. Lu MH, Tang B, Zeng S, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35(27):3524–3534. doi:10.1038/onc.2015.413
28. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi:10.1016/j.cell.2011.09.028
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.