Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA H19 Promotes Proliferation, Migration and Invasion and Inhibits Apoptosis of Breast Cancer Cells by Targeting miR-491-5p/ZNF703 Axis
Authors Wang Y, Wu Z, Li Y, Zheng Z, Yan J, Tian S, Han L
Received 14 January 2020
Accepted for publication 25 August 2020
Published 28 September 2020 Volume 2020:12 Pages 9247—9258
DOI https://doi.org/10.2147/CMAR.S246009
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Yongkun Wang,1,* Zhen Wu,1,* Yingxue Li,2 Zheng Zheng,2 Jinqiang Yan,2 Shuyan Tian,2 Lin Han2
1Department of Thyroid Surgery, Liaocheng People’s Hospital (Clinical Hospital of Shandong First Medical University) Liaocheng, Shandong, People’s Republic of China; 2Department of Pathology, Liaocheng People’s Hospital (Clinical Hospital of Shandong First Medical University) Liaocheng, Shandong, People’s Republic of China
*These authors contributed equally to this work.
Correspondence: Lin Han Tel + 86-0635-8273132
Email [email protected]
Background: Breast cancer is one of the most common cancers worldwide. Long non-coding RNAs and microRNAs act as important regulators in human cancers. This study aims to explore the molecular mechanism among H19, miR-491-5p and zinc finger 703 (ZNF703) in breast cancer.
Materials and Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to detect the expression of H19, miR-491-5p and ZNF703. Cell Counting Kit 8 (CCK-8) assay was performed to evaluate cell proliferation. Cell apoptosis was assessed by flow cytometry assay. The number of migrated and invaded cells was counted by transwell assay. Dual luciferase reporter assay was carried out to test luciferase activity. Protein level of ZNF703 was measured by Western blot assay.
Results: H19 was highly expressed in breast tissues and cells. H19 knockdown inhibited proliferation, induced apoptosis and blocked migration and invasion. Moreover, H19 bound to miR-491-5p and negatively regulated miR-491-5p expression. MiR-491-5p inhibition abrogated the activities of proliferation, apoptosis, migration and invasion affected by H19 knockdown. Furthermore, miR-491-5p directly targeted ZNF703 and inversely modulated ZNF703 expression. ZNF703 up-regulation rescued the effects of miR-491-5p overexpression on proliferation, apoptosis, migration and invasion. In addition, H19 knockdown reduced ZNF703 expression by targeting miR-491-5p/ZNF703 axis.
Conclusion: H19 promoted proliferation, migration and invasion and retarded apoptosis of breast cancer cells via sponging miR-491-5p to down-regulate ZNF703 expression.
Keywords: breast cancer, H19, miR-491-5p, ZNF703
Introduction
Breast cancer is often diagnosed in female and has developed into the second leading cause of cancer death worldwide recently.1 In 2018, over 2 million people were newly diagnosed with breast cancer which leads to more than 600,000 deaths every year.2 Numerous studies have revealed that the increasing incidence of breast cancer was due to many risk factors including obesity, alcohol abuse and genetic mutations.3 Although, a large of improvement has been made in the therapeutic methods and strategies of breast cancer, the overall survival is still unsatisfactory low.4 The activation of some signaling pathways related to cell proliferation also facilitated cell aberrant progression, resulting in the high recurrence of breast cancer.5 Hence, the knowledge about the pathogenesis of breast cancer is urgently needed for finding better therapies for breast cancer patients.
Long non-coding RNAs (lncRNAs) are a class of large RNA transcripts with more than 200 nts in length, which were originally regarded as “transcriptional noise” during the gene transcription process.6 The frequent dysregulation of lncRNAs has been reported in human cancers in recent years. They played a vital role in the carcinogenesis and cancer progression, also, some lncRNAs acted as important biomarkers for diagnosis and prognosis of human cancers, including breast cancer.7,8 H19 is transcribed from H19 gene that is located at the region of chromosome 11p15.9 Increasing studies demonstrated that H19 played a positive role in the embryonic development and tumorigenesis processes in multiple cancers. For example, zhang et al discovered that H19 expression was increased and H19 knockdown largely inhibited the progression of anaplastic thyroid carcinoma cells;10 also, H19 significantly enhanced the malignant development of HCC cells by targeting miR-22.11 Moreover, H19 has been identified to associate with the cell progression, cell chemosensitivity, poor prognosis and cell metastasis.12–15 However, more researches need to be explored for better understanding the molecular mechanism of H19 in breast cancer.
MicroRNAs (miRNAs), a large set of non-coding RNAs of 21–24 nucleotides, have focused many researchers’ attention from 1993 when the first miRNA was discovered in Caenorhabditis Elegans. MiRNAs acted as important regulators during the gene translation process or some malignant progression.16 MiRNA-491-5p (miR-491-5p) is located at the site of chromosome 9p21.3 and has been proved as tumor suppressor in various cancers such as lung cancer,17 colorectal cancer,18 gastric cancer,19 prostate cancer.20 Furthermore, miR-491-5p up-regulation prominently blocked the proliferation of breast cancer cells by targeting JMJD2B.21 Although some of the functions of miR-491-5p have been studied, the regulatory mechanism of H19/miR-491-5p axis is still unclear.
Zinc finger 703 (ZNF703) belongs to NocA/Nlz, Elbow, Tlp-1 (NET) family which is highly conserved transcriptional repressor.22 Several studies have showed that ZNF703 was involved in tumorigenesis and prognosis of diverse cancers.23,24 Also, Shi et al disclosed that ZNF703 was up-regulated and played an oncogenic role in breast cancer.25 Besides, Marzbany et al revealed that the silencing of ZNF703 obviously induced apoptosis of breast cancer cells combined with ibuprofen, which provided a combinatorial therapy for breast cancer.26 Despite so many researches of ZNF703 in breast cancer, the interaction among H19, miR-491-5p and ZNF703 has not been illuminated.
In our research, we detected the levels of H19, miR-491-5p and ZNF703 and elucidated the relationship between H19 and miR-491-5p, miR-491-5p and ZNF703. Also, we studied the effects of H19/miR-491-5p/ZNF703 axis on the progression of breast cancer.
Materials and Methods
Patients and Cell Culture
The tumor tissues and adjacent normal tissues were obtained from the breast cancer patients (n=20) who were diagnosed during March to September of 2018 in the hospital of Liaocheng People’s Hospital (Clinical Hospital of Shandong First Medical University). Any other treatment was not conducted on the breast cancer patients before this study. The written informed consent has obtained from every breast cancer patient. The Ethics committee of Liaocheng People’s Hospital (Clinical Hospital of Shandong First Medical University) has agreed all experiments in this study.
The normal human breast cell line MCF-10A and the breast cancer cell lines (BT-474, DU4475 and HCC1806) were purchased from American Tissue Culture Collection (ATCC, Manassas, VA, USA), and the other breast cancer cell line BT-549 was purchased from the Lombardi Cancer Center of Georgetown University (Washington, DC, USA). Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Rockford, IL, USA) was prepared in the plates to provide the nutrition for cell growth and 10% fetal bovine serum (FBS, Thermo Fisher Scientific) was also added into the medium. All cells were cultivated in incubator at 37°C.
Cell Transfection
The interference for H19 (si-H19) and its matched control (si-NC), the overexpression of miR-491-5p (miR-491-5p) and the blank control (miR-NC), the antisense for miR-491-5p (anti-miR-491-5p) and negative control (anti-miR-NC) were all provided by Sangon Biotech (Sangon, Shanghai, China). The overexpression vector of ZNF703 (ZNF703) and the empty vector (Vector) (GenePharma, Shanghai, China) were constructed. The Lipofectamine 2000 reagent was obtained from Thermo Fisher Scientific (Rockford, IL, USA) for the transfection of all above oligonucleotides and vectors.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The tissues from breast cancer patients and cell lines were all collected after resection surgery or cell cultivation. The extraction of total RNA was performed by lysing them in the Trizol reagent (Takara, Dalian, China). Then, Nanodrop 2000 (Thermo Fisher Scientific) and gel electrophoresis (Sigma, St. Louis, MO, USA) were conducted to test the quality and quantity of total RNA. Next, RT-PCR kit (Invitrogen, Carlsbad, CA, USA) was used for reverse transcription. The quantitative experiment was carried out with qPCR SuperMix (Transgen biotech). QRT-PCR results were standardized by the 2−ΔΔCt calculation method with the internal reference: U6 or Glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers were synthesized by Sangon Biotech (Sangon) and the sequences of H19, miR-491-5p, ZNF703, GAPDH and U6 primers were listed as follows: H19, 5ʹ-ATCGGTGCCTCAGCGTTCGG-3ʹ (Forward), 5ʹ-CTGTCCTCGCCGTCACACCG-3ʹ (Reverse); miR-491-5p, 5ʹ-GGAGTGGGGAACCCTTCC-3ʹ (Forward), 5ʹ-GTGCAGGGTCCGAGGT-3ʹ (Reverse); ZNF703, 5ʹ-AACGGCCCACATGAGTCAAT-3ʹ (Forward), 5ʹ-GGCGGGGATCATGTCGTTAT-3ʹ (Reverse); GAPDH, 5ʹ-ACAACAGCCTCAAGATCATCAG-3ʹ (Forward), 5ʹ-GGTCCACCACTGACACGTTG-3ʹ (Reverse); U6, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ (Forward), 5ʹ-GGAACGCTTCACGAATTTG-3ʹ (Reverse).
Cell Counting Kit 8 (CCK-8) Assay
The proliferation of BT-549 and BT-474 cells was evaluated by using CCK-8 (ACMEC, Shanghai, China). After cultivation for the indicate time (24 h, 48 h, 72 h), the transfected BT-549 and BT-474 cells were collected and digested with trypsin. 10 μL CCK-8 solution was placed to the transfected breast cancer cells for another 4 h cultivation. Then, the microplate reader (Bio-Rad, Hercules, CA, USA) was employed to measure the optical density at 450 nm.
Flow Cytometry
After transfection experiment, cells were harvested and suspended with 1 × binding buffer. Then, the Annexin V-fluorescein isothiocyanate (V-FITC)/propidium iodide (PI) Apoptosis Detection Kit (Thermo Fisher Scientific) was used to stain cells. Finally, the flow cytometer was used to detect the apoptosis rate of transfected BT-549 and BT-474 cells.
Transwell Assay
At the 48 h post-transfection, BT-549 and BT-474 cells were harvested and suspended in the serum-free medium. For the detection of cell invasion, Matrigel (Corning, New York, NY, USA) was performed to cover the upper transwell chamber (Corning), while for cell migration assessment, the chambers did not need any treatment. The transfected BT-549 and BT-474 cells were seeded into the upper chamber and followed by 12 h culturing in incubator. Subsequently, cells were incubated with 0.1% crystal violet for another 20 min and then cells on the upper layer of chamber were removed. Lastly, the inverted optical microscope was employed to detect the number of cells with three random fields.
Dual Luciferase Reporter Assay
The binding sites between H19 and miR-491-5p, between miR-491-5p and ZNF703 were predicted by starBase or TargetScan website tool, respectively. First, the wild type of H19 and ZNF703 were cloned and inserted into the psi-CHECK-2 vector (Promega, Madison, WI, USA), named as H19-WT and ZNF703-WT. The mutated sequences of H19 and ZNF703 were designed and synthesized by Sangon Biotech (Sangon), and then cloned into luciferase vector, named as H19-Mut and ZNF703-Mut. Each vector was transfected into breast cancer cells with miR-491-5p or miR-NC according to the cell transfection protocol. The luciferase activities were detected by using the luciferase reporter assay kit (Promega).
Western Blot Assay
Total proteins extraction was performed based on the instructions. After measurement of protein quality and quantity, proteins were loaded into freshly prepared sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Following 2 h electrophoresis at 100 v, the proteins were blotted onto the membranes (Thermo Fisher Scientific). After blocked for 1 h and rinsed by washing buffer for three times, the membranes were mixed with antibodies I against ZNF703 (ab188031; 1/500; Abcam, Cambridge, MA, USA) or β-actin (ab8227; 1/1000; Abcam). Then, they were mixed with antibody II (ab6721; 1/2000; Abcam) for another 2 h. The proteins on the membrane were treated with commercial enhanced chemiluminescence chromogenic substrate (Beyotime) and the expression of ZNF703 was analyzed by using Image Lab software (Bio-Rad) with normalization to that of β-actin internal control.
Statistical Analysis
All data were analyzed by SPSS 21.0 software and all results were shown as mean ± standard deviation (SD). The Student’s t-test was employed for data comparison between two groups and one-way analysis of variance (ANOVA) for comparisons among at least three groups. P < 0.05 was regarded to be statistically significant difference.
Results
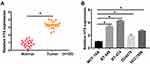
H19 Expression Was Sharply Increased in Breast Cancer Tissues and Cells
Firstly, qRT-PCR assay was conducted to detect the expression of H19 in tumor tissues and cells. The results showed that H19 was highly expressed in the tumor tissues compared with the normal tissues (Figure 1A). Also, H19 expression was greatly enhanced in breast cancer cell lines (BT-549, BT-474, DU4475 and HCC1806, especially in BT-549 and BT-474 cell lines) relative to normal breast cell line MCF-10A cell (Figure 1B), so BT-549 and BT-474 cell lines were selected for subsequent experiments. These data implied that H19 might be involved in the development of breast cancer.
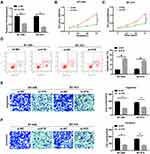
Silenced H19 Inhibited Proliferation, Migration and Invasion and Promoted Apoptosis of Breast Cancer Cells
To explore the role of H19 in breast cancer, BT-549 and BT-474 cells were transfected with si-H19 or si-NC. QRT-PCR assay indicated that the expression of H19 was significantly decreased in BT-549 and BT-474 cells transfected with si-H19 (Figure 2A). Moreover, CCK-8 assay showed that the proliferation of BT-549 and BT-474 cells was markedly blocked by the down-regulation of H19 (Figure 2B and C). Also, the apoptosis rate was examined by flow cytometry assay and the results implicated that the apoptosis of BT-549 and BT-474 cells was induced by the transfection with si-H19 (Figure 2D). Transwell assay determined that both migration and invasion of BT-549 and BT-474 cells were hindered by the silencing of H19 (Figure 2E and F). All these data revealed that H19 silencing inhibited the proliferation, migration and invasion and triggered apoptosis of breast cancer cells.
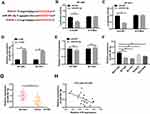
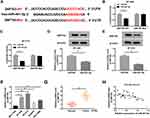
H19 Directly Bound to miR-491-5p and Negatively Regulated miR-491-5p Expression
To elucidate the relationship between H19 and miR-491-5p, starBase online tool was conducted to predict the binding site between them. The putative binding sequences among H19-WT, miR-491-5p and H19-Mut are shown in Figure 3A. Next, dual luciferase reporter assay was performed to confirm the prediction. The results demonstrated that miR-491-5p co-transfection with H19-WT limited the luciferase activities of BT-549 and BT-474 cells compared with negative control, while there was no change of luciferase activities when H19-Mut co-transfection with miR-491 or miR-NC (Figure 3B and C). Also, qRT-PCR assay was conducted in breast cancer cells and revealed that miR-491-5p expression was prominently elevated by H19 knockdown in BT-549 and BT-474 cells (Figure 3D). Nevertheless, miR-491-5p could not affect the expression of H19 (Figure 3E). Meanwhile, the expression of miR-491-5p was largely promoted in BT-549, BT-474, DU4475 and HCC1806 cells compared with MCF-10A cells (Figure 3F). In addition, we detected the expression of miR-491-5p in patient tissues, and the results showed that miR-491-5p was down-regulated in breast cancer tissues (Figure 3G). Besides, the level of miR-491-5p was negatively correlated with H19 level in breast cancer tissues (Figure 3H). In total, H19 reduced miR-491-5p expression by directly targeting miR-491-5p.
MiR-491-5p Interference Recovered H19 Knockdown Mediated Anti-Proliferation, Pro-Apoptosis, Anti-Migration and Anti-Invasion
To illuminate the interaction between H19 and miR-491-5p in breast cancers, BT-549 and BT-474 cells were transfected with si-NC, si-H19, si-H19 + anti-miR-NC or si-H19 + anti-miR-491-5p. CCK-8 assay disclosed that H19 silencing inhibited the proliferation of BT-549 and BT-474 cells, which was abated by transfection with miR-491-5p inhibition (Figure 4A and B). Flow cytometry assay showed that the apoptosis was promoted by H19 knockdown and then impeded after co-transfection with miR-491-5p inhibition (Figure 4C and D). Furthermore, miR-491-5p inhibition reversed the inhibitory effect of H19 silencing on migration and invasion of BT-549 and BT-474 cells (Figure 4E and F). In a word, H19 knockdown restrained proliferation, migration and invasion and induced apoptosis of breast cancer cells by regulating miR-491-5p.
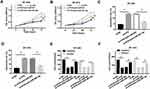
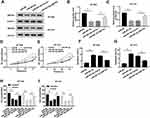
ZNF703 Was a Target of miR-491-5p and ZNF703 Expression Was Suppressed by miR-491-5p
To search for the downstream targets of miR-491-5p, TargetScan website tool was performed to predict the target mRNAs. Interestingly, ZNF703 has the binding sites with miR-491-5p. The potential binding sequences between miR-491-5p and ZNF703 and the mutant sequences of ZNF703 were exhibited in Figure 5A. To verify the prediction, dual luciferase reporter assay was conducted in BT-549 and BT-474 cells co-transfection with ZNF703-WT or ZNF703-Mut and miR-NC or miR-491-3p. The results manifested that luciferase activities of BT-549 and BT-474 cells were drastically reduced by miR-491-5p compared with negative control in ZNF703-WT group, while no obvious change of luciferase activities was observed in ZNF703-Mut group (Figure 5B and C). Also, Western blot assay showed that miR-491-5p overexpression notably repressed ZNF703 protein level in BT-549 and BT-474 cells (Figure 5D and E). Moreover, the mRNA level of ZNF703 was higher in BT-549, BT-474, DU4475 and HCC1806 cells than that in MCF-10A cells (Figure 5F). Additionally, ZNF703 mRNA level was up-regulated in breast cancer tissues and negatively correlated with miR-491-5p (Figure 5G and H). These data suggested that miR-491-5p targeted ZNF703 and suppressed ZNF703 level in breast cancer cells.
ZNF703 Mitigated the Activities of miR-491-5p Overexpression on Proliferation, Apoptosis Migration and Invasion
To further explore the relation between miR-491-5p and ZNF703 in breast cancer, the protein level of ZNF703 was detected in cells transfected with miR-491-5p. The results indicated that miR-491-5p overexpression triggered a significant reduction of ZNF703 level in BT-549 and BT-474 cells, however, ZNF703 level was then enhanced after co-transfection with ZNF703 (Figure 6A and C). Moreover, CCK-8 assay revealed that miR-491-5p repressed the proliferation of BT-549 and BT-474 cells, which was rescued by introduction with ZNF703 (Figure 6D and E). Flow cytometry assay displayed that miR-491-5p largely promoted the apoptosis of BT-549 and BT-474 cells, while cell apoptosis was hindered by ZNF703 up-regulation (Figure 6F and G). Furthermore, transwell assay was performed to measure the migration and invasion and disclosed that both migrated and invaded cells were reduced by miR-491-5p overexpression, and they were increased when co-transfection with miR-491-5p and ZNF703 (Figure 6H and I). In addition, ZNF703 knockdown inhibited proliferation, migration and invasion and induced apoptosis of BT-549 and BT-474 cells (Supplementary Figure 1). Collectively, miR-491-5p repressed proliferation, migration and invasion and enhanced apoptosis of breast cancer cells by regulating ZNF703.
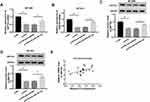
H19 Knockdown Reduced ZNF703 Expression by Sponging miR-491-5p
To illuminate the impact of H19/miR-491-5p axis on ZNF704 expression, protein and mRNA levels of ZNF703 were detected by qRT-PCR or Western blot assays, respectively. The results revealed that both protein and mRNA levels of ZNF703 were dramatically decreased by silenced H19 in BT-549 and BT-474 cells, which was attenuated following transfection with miR-491-5p inhibition (Figure 7A and D). Besides, there is a positive correlation between H19 and ZNF703 mRNA levels in breast cancer tissues (Figure 7E). Totally, H19 silencing reduced ZNF703 level through targeting miR-491-5p.
Discussion
Breast cancer is a complex malignancy which caused by a variety of genetic changes.27 It has been widely reported that plenty of lncRNAs and miRNAs were involved in tumorigenesis and the development of breast cancer. Despite some advances have been made to improve the overall survival, it is urgent to develop a possible and effective treatment for breast cancer.
Wang et al discovered that H19 is up-regulated and induced cell autophagy resulting in the tamoxifen resistance in breast cancer.28 Further, H19 expression was up-regulated in the plasma of breast cancer patients and H19 inhibition drastically repressed cell proliferation.29 Consistent with above studies, we investigated the expression and the role of H19 in breast cancer. The results indicated that H19 level was elevated in breast tumor tissues. The silencing of H19 significantly retarded the cell proliferation, metastasis and induced apoptosis. These data suggested that H19 knockdown limited the cell growth in breast cancer.
A previous report indicated that miR-491-5p acted as a tumor suppressor in breast cancer,21 and miR-491-5p could affect the cancer susceptibility by binding MMP9.30 In order to illuminate the relationship between H19 and miR-491-5p, we applied the starBase online tool to predict the binding site between them. Meanwhile, the results of dual luciferase assay verified this combination and miR-491-5p expression was enhanced by H19 down-regulation. Moreover, the proliferation, migration and invasion were inhibited by H19 knockdown and then were reversed by miR-491-5p inhibition. On the contrary, H19 knockdown induced the apoptosis which was restrained by miR-491-5p inhibition. These data demonstrated that H19 knockdown hampered the progression of breast cancer cells by suppressing miR-491-5p expression.
TargetScan tool was used to predict the potential target of miR-491-5p and we found that ZNF703 has the imperfect combination possibility with miR-491-5p. Interestingly, Marzbany et al disclosed that there was an increased expression of ZNF703 in breast cancer tissues and cells.26 Then, we conducted dual luciferase reporter assay to prove the prediction, and Western blot assay implicated that the level of ZNF703 was negatively regulated by miR-491-5p. Furthermore, ZNF703 rescued the activities of miR-491-5p overexpression on proliferation, apoptosis, migration and invasion. Furthermore, the level of ZNF703 was decreased by H19 knockdown via sponging miR-491-5p. These results suggested that H19 contributed to the progression of breast cancer cells via targeting miR-491-5p to up-regulate ZNF703 expression.
This study revealed the functions of H19 in breast cancer and the molecular mechanism of H19/miR-491-5p/ZNF703 axis in vitro, while the in vivo data from the mice model or breast cancer patients remain to study further.
Conclusions
The level of H19 was increased in breast tumor tissues and cells. H19 silencing hindered the breast cancer cells proliferation and metastasis and triggered apoptosis. Also, H19 bound to miR-491-5p and inversely regulated miR-491-5p expression. H19 knockdown impeded breast cancer cell progression through targeting miR-491-5p. Moreover, ZNF703 was targeted by miR-491-5p and ZNF703 expression was decreased by miR-491-5p. MiR-491-5p repressed the progression of breast cancer cells via down-regulating ZNF703 expression. Besides, ZNF703 level was modulated by H19/miR-491-5p axis. These data provided new evidence for better study of the role of H19 and might provide an effective target for the treatment of breast cancer.
Highlights
Although the role of H19 has been reported before, the mechanism of H19 regulating miR-491-5p was unknown.
MiR-491-5p was proved to act a tumor suppressor in breast cancer by targeting JMJD2B, while the interaction between miR-491-5p and ZNF703 remained unclear.
The molecular mechanism of H19/miR-491-5p/ZNF703 in breast cancer has not been explored before.
Abbreviations
ZNF703, zinc finger 703; qRT-PCR, quantitative real-time polymerase chain reaction; CCK-8, Cell counting Kit 8; lncRNAs, long non-coding RNAs; ATCC, American Tissue Culture Collection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; V-FITC, V-fluorescein isothiocyanate; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SD, standard deviation; ANOVA, analysis of variance.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of The Liaocheng People’s Hospital (Clinical Hospital of Shandong First Medical University).
Patient Consent for Publication
Not applicable.
Author Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Funding
No funding was received.
Disclosure
Yongkun Wang and Zhen Wu are co-first authors for this study. The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Rudolph A, Chang-Claude J, Schmidt MK. Gene-environment interaction and risk of breast cancer. Br J Cancer. 2016;114(2):125–133. doi:10.1038/bjc.2015.439
4. Dai K, Qin F, Zhang H, et al. Low expression of BMPRIB indicates poor prognosis of breast cancer and is insensitive to taxane-anthracycline chemotherapy. Oncotarget. 2016;7(4):4770–4784. doi:10.18632/oncotarget.6613
5. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674.
6. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
7. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208.
8. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64.
9. Delaval K, Wagschal A, Feil R. Epigenetic deregulation of imprinting in congenital diseases of aberrant growth. Bioessays. 2006;28(5):453–459. doi:10.1002/bies.20407
10. Zhang H, Yu Y, Zhang K, et al. Targeted inhibition of long non-coding RNA H19 blocks anaplastic thyroid carcinoma growth and metastasis. Bioengineered. 2019;10(1):306–315. doi:10.1080/21655979.2019.1642722
11. Li L, Han T, Liu K, et al. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci. 2019;23(12):5392–5401.
12. Si H, Chen P, Li H, Wang X. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am J Transl Res. 2019;11(5):3213–3225.
13. Gao H, Hao G, Sun Y, et al. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001–8012. doi:10.2147/OTT.S172379
14. Shima H, Kida K, Adachi S, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170(3):507–516. doi:10.1007/s10549-018-4793-z
15. Yoshimura H, Matsuda Y, Yamamoto M, et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab Invest. 2018;98(6):814–824. doi:10.1038/s41374-018-0048-1
16. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi:10.1016/j.ydbio.2006.08.028
17. Chen Y, Zhang Y, Song W, et al. Ginsenoside Rh2 inhibits migration of lung cancer cells under hypoxia via mir-491. Anticancer Agents Med Chem. 2019;19:1633–1641. doi:10.2174/1871520619666190704165205
18. Lu L, Cai M, Peng M, et al. miR-491-5p functions as a tumor suppressor by targeting IGF2 in colorectal cancer. Cancer Manag Res. 2019;11:1805–1816. doi:10.2147/CMAR.S183085
19. Yu T, Wang LN, Li W, et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109(5):1393–1403. doi:10.1111/cas.13583
20. Xu Y, Hou R, Lu Q, et al. MiR-491-5p negatively regulates cell proliferation and motility by targeting PDGFRA in prostate cancer. Am J Cancer Res. 2017;7(12):2545–2553.
21. Hui Z, Yiling C, Wenting Y, et al. miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERalpha-positive breast cancer. FEBS Lett. 2015;589(7):812–821. doi:10.1016/j.febslet.2015.02.014
22. Runko AP, Sagerstrom CG. Nlz belongs to a family of zinc-finger-containing repressors and controls segmental gene expression in the zebrafish hindbrain. Dev Biol. 2003;262(2):254–267. doi:10.1016/S0012-1606(03)00388-9
23. Yang G, Ma F, Zhong M, et al. ZNF703 acts as an oncogene that promotes progression in gastric cancer. Oncol Rep. 2014;31(4):1877–1882. doi:10.3892/or.2014.2997
24. Li K, Wang J, Han J, et al. Overexpression of ZNF703 facilitates tumorigenesis and predicts unfavorable prognosis in patients with cholangiocarcinoma. Oncotarget. 2016;7(46):76108–76117. doi:10.18632/oncotarget.12627
25. Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:
26. Marzbany M, Bishayee A, Rasekhian M. Increased expression of ZNF 703 in breast cancer tissue: an opportunity for RNAi-NSAID combinatorial therapy. Biotechnol Appl Biochem. 2019;66:808–814. doi:10.1002/bab.1790
27. Motamedolshariati M, Memar B, Aliakbaian M, et al. Accuracy of prognostic and predictive markers in core needle breast biopsies compared with excisional specimens. Breast Care. 2014;9(2):107–110. doi:10.1159/000360787
28. Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12(1):81. doi:10.1186/s13045-019-0747-0
29. Muller V, Oliveira-Ferrer L, Steinbach B, et al. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 2019;13(5):1137–1149. doi:10.1002/1878-0261.12472
30. Pirooz HJ, Jafari N, Rastegari M, et al. Functional SNP in microRNA-491-5p binding site of MMP9 3ʹ-UTR affects cancer susceptibility. J Cell Biochem. 2018;119(7):5126–5134. doi:10.1002/jcb.26471
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.