Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Cell Progression and Paclitaxel Resistance by Regulating miR-126-5p/ATAD2 Axis
Authors Liu C, Hou J, Shan F, Wang L, Lu H, Ren T
Received 6 November 2019
Accepted for publication 21 April 2020
Published 2 June 2020 Volume 2020:13 Pages 4931—4942
DOI https://doi.org/10.2147/OTT.S237580
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Chang Liu, Jianfeng Hou, Fengxiao Shan, Lijuan Wang, Hanjie Lu, Tiejun Ren
Department of Oncology, Luoyang Central Hospital Affiliated to Zhengzhou University, Luoyang 471000, People’s Republic of China
Correspondence: Chang Liu; Tiejun Ren
Department of Oncology, Luoyang Central Hospital Affiliated to Zhengzhou University, No. 288, Zhongzhou Road, Luoyang, Henan 471000, People’s Republic of China
Email [email protected]; [email protected]
Background: Long non-coding RNA colorectal neoplasia differentially expressed (lncRNA CRNDE) and microRNA-126-5p (miR-126-5p) were reported to be related to the development of colorectal carcinoma (CRC). However, the detailed mechanism of CRNDE and miR-126-5p is not fully understood. The purpose of this research was to explore their roles and molecular mechanism in CRC.
Methods: Quantitative real-time polymerase chain reaction was performed to detect the transcription levels of genes. Paclitaxel (PTX) was used to analyze cell drug resistance. 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay and flow cytometry analysis were employed to assess cell proliferation and apoptosis, respectively. Furthermore, cell migratory and invasive abilities were measured using transwell assay. The interaction between miR-126-5p and CRNDE or ATPase family AAA domain-containing protein 2 (ATAD2) was predicted by online tool starbase and then confirmed using the dual-luciferase reporter assay. Besides, Western blot assay was carried out to detect the levels of proteins.
Results: CRNDE and ATAD2 expressions were upregulated and miR-126-5p expression was downregulated in CRC tissues and cells. CRNDE depletion repressed PTX resistance and the growth of CRC cells. Interestingly, we found that miR-126-5p was a target gene of CRNDE, and miR-126-5p directly targeted ATAD2. Furthermore, CRNDE affected CRC cell progression via modulation of miR-126-5p/ATAD2 axis in CRC cells.
Conclusion: Our data suggested that CRNDE regulated CRC cell development and PTX resistance by modulating miR-126-5p/ATAD2 axis, providing the theoretical basis for the treatment of CRC patients.
Keywords: CRNDE, miR-126-5p, ATAD2, PTX resistance, cell progression, colorectal carcinoma
Introduction
Colorectal carcinoma (CRC), the third popular tumor, has a high recurrence rate and seriously threatens people’s health around the world.1,2 According to the statistic in 2015, there were approximately 1,400,000 new cases and an estimated 690,000 deaths of CRC every year.3 Nowadays, chemotherapy is an important strategy for the treatment of CRC patients, and many drugs, such as platinum, methotrexate (MTX), and paclitaxel (PTX), are widely applied.4 However, drug resistance is the main obstacle to the development of chemotherapy. Therefore, it is essential to explore the mechanism of CRC development for the therapy of CRC patients.
Long non-coding RNAs (lncRNAs), with approximately 200 nucleotides, are a group of conserved RNAs that play crucial roles through regulating gene expression or epigenetics and are considered as biomarkers for the prognosis and diagnosis of human cancers.5–7 LncRNA Colorectal Neoplasia Differentially Expressed (CRNDE), a gene located on chromosome 16, was highly expressed in CRC.8 Moreover, increased CRNDE expression was observed in many other human cancers, including ovarian cancer,9 glioma,10 cervical cancer,11 and non-small cell lung cancer.12 These data revealed that CRNDE was related to the development of human cancers. In recent years, some papers suggested that CRNDE positively regulated the growth of CRC cells. For example, Han et al confirmed that CRNDE induced cell proliferation and chemotherapy through regulating miR-181a-5p expression in CRC cells.13 Therefore, the studies of CRNDE in CRC are important.
MicroRNAs (miRNAs) are small non-coding RNAs that have approximately 22 nucleotides and regulate various cell behaviors, including proliferation, invasion, and apoptosis, through modulating the expressions of downstream genes.14,15 During recent years, miRNAs have been reported as a class of regulators to regulate CRC development. For example, Schetter et al suggested that miRNA expression was altered in CRC and related to cell growth and metastasis.16 Slaby et al demonstrated that miRNAs exerted function in the progression of CRC development and had an implicated expression pattern in CRC tissues.17 As a miRNA, miR-126-5p was reported as a regulator that was related to the development of CRC.18 However, the detailed mechanism of miR-126-5p in CRC is not fully understood.
ATPase family AAA domain-containing protein 2 (ATAD2) contains two domains, AAA+ domain regulates the function of substrate protein via affecting its conformation, and bromodomain interacts with acetylated lysine of substrate protein.19–21 Present evidence suggested that ATAD2 was related to a variety of human cancers. For example, ATAD2 level was increased and ATAD2 knockdown repressed angiogenesis in retinoblastoma.22 Ji et al confirmed that the silence of ATAD2 repressed cell migration and invasion in renal cell carcinoma.23 In CRC, it was reported that ATAD2 expression was increased and high ATAD2 level associated with a short overall survival of CRC patients.24 Therefore, the studies of ATAD2 in CRC are needed.
Here, we first detected CRNDE, miR-126-5p, and ATAD2 expressions in CRC tissues and cells, and then investigated the function of CRNDE in cell proliferation, migration, invasion, apoptosis, and PTX resistance of CRC cells. Furthermore, the functions of miR-126-5p and ATAD2 in CRNDE-regulated cell growth were explored in CRC cells.
Materials and Methods
Tissue Samples and Cell Culture
CRC tissues and adjacent tissues were obtained from 41 patients with CRC who received surgical treatment (among them, there are 31 paired CRC tissues and adjacent tissues) and normal tissues were obtained from 41 patients without CRC who received surgical treatment at Luoyang Central Hospital Affiliated to Zhengzhou University. All tissues were stored at −80°C for further experiment. This assay was approved by the ethics committee of Luoyang Central Hospital Affiliated to Zhengzhou University. All patients signed informed consents and did not receive any therapy before this assay.
Human normal cell line (NCM460) was provided by Xiehe Cell Bank of the Chinese Academy of Medical Sciences (Beijing, China). CRC cell lines (Caco-2, HT29, HCT15, SW480, SW620) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; Gibco) with 5% CO2 at 37°C.
In this study, different concentrations of PTX (0, 0.166, 0.313, 0.625, 1.25, 2.5, 5, 10, or 20 µM/L) were used to analyze drug resistance of CRC cells.
Plasmid and Transfection
Si-CRNDE, miR-126-5p mimic (miR-126-5p), miR-126-5p inhibitor (anti-miR-126-5p), si-ATAD2 and their controls (si-NC, miR-NC, and anti-miR-NC) were obtained from Ribobio (Shanghai, China). CRNDE sequence was amplified and then cloned into the pcDNA3.1 plasmid (Genepharma, Shanghai, China) to generate CRNDE overexpression vector.
When the confluence of cells was up to 70%, cell transfection assay was carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) based on the user’s manual.
RNA Extraction and Quantitative Real-Time Polymerase Chain (qRT-PCR) Assay
Total RNA in CRC tissues or cells was obtained using Trizol reagent (Life Technologies Corporation, Carlsbad, CA, USA) based on the user’s manual. Then, superscript II first-strand synthesis system (Invitrogen) was used to generate cDNA, and qRT-PCR was carried out with SYBR green (Applied Biosystems, Foster City, CA, USA). The analysis of relative expression was conducted using the 2−ΔΔCt method with U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal control. The primers for CRNDE, miR-126-5p, and ATAD2, U6, and GAPDH were listed as follows: CRNDE (Forward, 5ʹ-ATATTCAGCCGTTGGTCTTTGA-3ʹ; Reverse, 5ʹ-TCTGCGTGACAACTGAGGATTT-3ʹ), miR-126-5p (Forward, 5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCGTA-3ʹ; Reverse, 5ʹ-ACACTCCAGCTGGGCATTATTACTTTTGGTA-3ʹ), ATAD2 (Forward, 5ʹ-GGAATCCCAAACCACTGGACA-3ʹ; Reverse, 5ʹ-GGTAGCGTCGTCGTAAAGCACA-3ʹ), U6 (Forward, 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ; Reverse, 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ), and GAPDH (Forward, 5ʹ-ATTCCATGGCACCGTCAAGGCTGA-3ʹ; Reverse, 5ʹ-TTCTCCATGGTGGTGAAGACGCCA-3ʹ).
Cell Proliferation Assay
In this study, the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was carried out to examine cell proliferation according to the recommended protocol (Promega, Madison, WI, USA). In brief, transfected SW480 and SW620 cells (5×103 cells/well) were cultured for 0 h, 24 h, 48 h, or 72 h, respectively. For drug resistance detection, transfected cells were cultured for 48 h and then treated with PTX for 24 h. Then, 20 µL MTT solution was applied to treat the cells for 4 h. Next, 100 µL dimethyl sulfoxide (DMSO) was used for dissolution of the formazan product. Finally, the absorbance of every sample was examined at 490 nm through the use of the microplate reader (Bio-Rad, Richmond, CA, USA).
Cell Apoptosis Assay
In this research, cell apoptosis rate was analyzed using Annexin V-Fluorescein (FITC)/PI apoptosis detection kit (Solarbio, Beijing, China) and flow cytometry (BD Biosciences, San Jose, CA, USA) in line with the recommended instruction. Briefly, after transfection for 48 h, SW480 and SW620 cells (2×105 cells) were harvested, and then stained with 5 μL Annexin V-FITC and PI for 10 min under the dark condition, subsequently, the flow cytometry was applied to examine cell apoptosis.
Cell Migration and Invasion Assay
Cell migratory and invasive abilities were measured by a transwell chamber (Millipore, Billerica, MA, USA). Matrigel was coated on the upper chamber for the analysis of cell invasion. Briefly, after transfection for 48 h, SW480 and SW620 cells (5×104 cells) were harvested, mixed with serum-free DMEM medium (100 µL), and then transferred into the upper chamber. A 500 µL relative medium containing 10% FBS was seeded to the lower chamber. After culturing at 37°C for 12 h, the migratory or invasive cells were fixed with paraformaldehyde, stained by crystal violet, and counted under a microscope (Olympus, Tokyo, Japan).
The Dual-Luciferase Reporter Assay
For the constructions of WT/MUT-CRNDE and WT/MUT-ATAD2-3ʹuntranslated region (WT/MUT-ATAD2-3ʹUTR), corresponding sequences were amplified and then inserted into the pGL3 vector (Promega). Next, SW480 and SW620 cells were transfected with each of these reporter vectors and miR-126-5p or miR-NC. Finally, the Dual-Luciferase Reporter Assay System (Promega) was applied for the determination of the luciferase density after the cells were cultured for 48 h.
Western Blot Assay
Firstly, the proteins were obtained from SW480 and SW620 cells lysed by RIPA lysis buffer (Beyotime, Shanghai, China). Next, the proteins were subjected to 10% SDS-PAGE and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). After blocked by 5% TBST containing 5% non-fat skim for 2 h, the membranes were incubated with the primary antibody against ATAD2 or GAPDH (1:1000; Abcam, Cambridge, MA, USA) at 4°C overnight, and then incubated by corresponding secondary antibody (1:400; Abcam) for 1 h. Finally, the ECL Western blot kit (Beyotime) was used for the measurement of protein bands.
Statistical Analysis
All data were presented as the mean ± standard deviation (SD) under at least three independent experiment conditions. Student’s t-test was used for the analysis of a significant difference. Spearman correlation coefficient was carried out to investigate the relationship between levels of two genes. P less than 0.05 was considered statistically significant.
Results
CRNDE Expression Is Increased in CRC Tissues and Cells
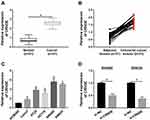
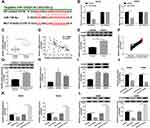
To investigate the role of CRNDE in CRC, we first detected the expression level of CRNDE in CRC tissues and normal tissues. The results suggested that CRNDE expression was significantly increased in CRC tissues (Figure 1A). Subsequently, we found that CRNDE level was higher in CRC tissues than that in adjacent tissues (Figure 1B). Besides, CRNDE expression in normal cells (NCM460) and CRC cells (Caco2, HT29, HCT15, SW480, and SW620) was also determined. As shown in Figure 1C, CRNDE expression was dramatically upregulated in CRC cells, especially in SW480 and SW620 cell lines, so these two cell lines were used for subsequent experiments. Therefore, CRNDE may exert function as a positive regulator in CRC development.
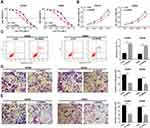
CRNDE Knockdown Inhibits Proliferation, Migration, Invasion and PTX Resistance, and Induces Apoptosis of CRC Cells
To further explore the function of CRNDE in CRC development, si-CRNDE was transfected into SW480 and SW620 cells to deplete CRNDE. Firstly, qRT-PCR analysis confirmed that the transfection with si-CRNDE strongly downregulated the expression level of CRNDE (Figure 1D). Next, different concentrations of PTX were used to treat the cells, and then the cell viability was assessed by MTT assay. The results demonstrated that cell viability was downregulated by CRNDE depletion (Figure 2A), meaning that CRNDE knockdown repressed PTX resistance of the cells. Besides, we determined the effect of CRNDE knockdown on 5-fluorouracil (5-FU) resistance, but the results showed that knockdown of CRNDE did not affect 5-FU resistance in SW480 and SW620 cells (Supplementary Figure 1). Meanwhile, MTT assay confirmed that CRNDE knockdown suppressed cell proliferation in SW480 and SW620 cells (Figure 2B). Also, trypan blue staining exhibited the same result with that of MTT assay (Supplementary Figure 2). In addition, flow cytometry analysis was performed to measure the cell apoptosis rate. As demonstrated in Figure 2C, cell apoptosis was dramatically induced by the depletion of CRNDE. Besides, decreased cell migratory ability (Figure 2D) and invasive ability (Figure 2E) were observed in CRNDE-depleted SW480 and SW620 cells. Moreover, the levels of migration and invasion-related proteins (E-cadherin, vimentin and MMP9) were detected, and the results showed that E-cadherin was upregulated and vimentin and MMP9 were downregulated by CRNDE knockdown, suggesting that knockdown of CRNDE repressed cell migration and invasion in CRC cells (Supplementary Figure 3). These data indicate that CRNDE knockdown inhibits cell growth and PTX resistance in CRC cells.
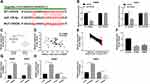
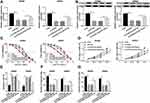
miR-126-5p Is a Target Gene of CRNDE
To understand the functional mechanism of CRNDE in CRC development, online tool starbase was used to predict the target genes of CRNDE. We found that miR-126-5p possessed complementary sequences with CRNDE (Figure 3A). Then, wide-type and mutant-type CRNDE luciferase reporter vectors (WT/MUT-CRNDE) were constructed and co-transfected into SW480 and SW620 cells with miR-NC or miR-126-5p. The analysis of luciferase activity suggested that the luciferase activity of WT-CRNDE was strongly repressed by miR-126-5p, whereas miR-126-5p did not affect the luciferase activity of MUT-CRNDE (Figure 3B). Thus, miR-126-5p interacted with CRNDE.
Next, miR-126-5p expression in CRC tissues and normal tissues was analyzed. As demonstrated in Figure 3C, miR-126-5p was lowly expressed in CRC tissues. Furthermore, we confirmed that miR-126-5p level was negatively correlated with CRNDE level in CRC tissues (Figure 3D). Besides, miR-126-5p expression was lower in CRC tissues than that in adjacent tissues (Figure 3E), and decreased miR-126-5p expression was also observed in CRC cells (Figure 3F). Finally, whether miR-126-5p expression was regulated by CRNDE was explored through transfecting si-CRNDE or CRNDE into SW480 and SW620 cells. We first confirmed that the transfection of CRNDE significantly increased CRNDE expression (Figure 3G), and then found that CRNDE knockdown dramatically upregulated miR-126-5p expression, as well as CRNDE overexpression remarkably downregulated miR-126-5p expression (Figure 3H). Therefore, CRNDE negatively regulates miR-126-5p expression through interaction in CRC cells.
CRNDE Regulates the Growth of CRC Cells Through Modulating miR-126-5p Expression
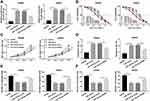
Based on the above results, it was speculated that CRNDE regulated miR-126-5p expression to modulate CRC cell growth. To verify this hypothesis, SW480 and SW620 cells were transfected with miR-NC, miR-126-5p, miR-126-5p + vector, or miR-126-5p + CRNDE, respectively. Firstly, qRT-PCR analysis suggested that miR-126-5p expression was upregulated by the transfection of miR-126-5p, and then partly rescued by the transfection of CRNDE (Figure 4A). Then, PTX resistance was analyzed by determining cell viability after PTX treatment. As shown in Figure 4B, the upregulation of miR-126-5p reduced PTX resistance, whereas this effect was weakened by CRNDE overexpression. Furthermore, we found that CRNDE overexpression induced cell proliferation suppressed by miR-126-5p upregulation in SW480 and SW620 cells (Figure 4C). Besides, the negative effect of CRNDE overexpression on miR-126-5p upregulation-regulated cell apoptosis was discovered in SW480 and SW620 cells (Figure 4D). Finally, cell migratory and invasive abilities were examined. As expected, the overexpression of CRNDE reversed the effect of miR-126-5p upregulation on CRC cell migration and invasion (Figure 4E and F). These data suggest that CRNDE mediates miR-126-5p expression to regulate cell progression in CRC cells.
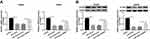
miR-126-5p Is a Molecular Sponge for ATAD2
In this study, ATAD2 was found as a potential target gene of miR-126-5p through using online tool starbase (Figure 5A). Then, the dual-luciferase reporter assay was carried out to verify this interaction. The results demonstrated that the luciferase activity of WT-ATAD2-3ʹUTR, but not MUT-ATAD2-3ʹUTR, was remarkably diminished by miR-126-5p (Figure 5B), confirming that miR-126-5p interacted with ATAD2. Next, qRT-PCR assay was performed to detect ATAD2 levels in CRC tissues and normal tissues. As shown in Figure 5C and D, ATAD2 level was increased in CRC tissues and negatively correlated with miR-126-5p level. Meanwhile, Western blot assay was carried out to determine ATAD2 protein level. Consistent with that in mRNA level, ATAD2 protein level was higher in CRC tissues than normal tissues (Figure 5E). Also, we found that the mRNA level and protein level of ATAD2 were significantly upregulated in CRC tissues (Figure 5F and G) and cells (Figure 5H and I) compared with that in adjacent tissues and normal cells.
Subsequently, the effect of miR-126-5p on ATAD2 expression was analyzed by transfecting miR-126-5p or anti-miR-126-5p into SW480 and SW620 cells. As shown in Figure 5J, the transfection of anti-miR-126-5p significantly downregulated miR-126-5p expression, verifying the efficiency of miR-126-5p knockdown. Then, qRT-PCR assay and Western blot assay demonstrated that the expression level of ATAD2 was downregulated by miR-126-5p overexpression and upregulated by miR-126-5p knockdown (Figure 5K and L). Taken together, miR-126-5p directly downregulates ATAD2 expression.
miR-126-5p Mediates ATAD2 Level to Affect CRC Cell Progression
Next, we investigated whether miR-126-5p mediating ATAD2 expression to regulate CRC cell growth via transfecting si-NC, si-ATAD2, si-ATAD2 + anti-miR-NC, or si-ATAD2 + anti-miR-126-5p into SW480 and SW620 cells. QRT-PCR analysis and Western blot assay confirmed that ATAD2 expression was downregulated by the transfection of si-ATAD2, and then partly rescued due to miR-126-5p knockdown (Figure 6A and B). Then, PTX resistance of the cells was assessed. As demonstrated in Figure 6C, miR-126-5p knockdown increased cell viability repressed by the downregulation of ATAD2 after PTX treatment. Moreover, the negative effect of miR-126-5p depletion on ATAD2 downregulation-regulated cell proliferation was observed in SW480 and SW620 cells (Figure 6D). Subsequently, flow cytometry analysis was used to analyze the cell apoptosis rate. The results suggested that the downregulation of miR-126-5p weakened the effect of ATAD2 knockdown on CRC cell apoptosis (Figure 6E). Besides, we found that the silence of ATAD2 repressed cell migration and invasion, whereas this action was reversed by decreased miR-126-5p expression (Figure 6F and G). Thus, miR-126-5p regulates CRC cell progression via modulation of ATAD2 level in CRC cells.
CRNDE Upregulates ATAD2 Level via Inhibiting miR-126-5p Expression
Finally, to determine whether CRNDE repressed miR-126-5p expression to increase ATAD2 level, SW480 and SW620 cells were transfected with miR-NC, miR-126-5p, miR-126-5p + vector, or miR-126-5p + CRNDE, respectively. Then, qRT-PCR assay and Western blot assay were conducted to detect the transcription level and protein level of ATAD2. The data indicated that the expression level of ATAD2 was downregulated by miR-126-5p overexpression, and then partly rescued by increased CRNDE expression (Figure 7A and B). Taken together, CRNDE inhibits miR-126-5p expression to upregulate the ATAD2 level in CRC cells.
Discussion
In the past few years, the dysregulation of lncRNAs was discovered in CRC and these lncRNAs played pivotal roles in CRC cell progression.25 For instance, Cui et al confirmed that lncRNA HEIH positively regulated the growth of CRC cells via modulating miR-939/Bcl-xL axis.26 Iguchi et al demonstrated that lncRNA ATB was remarkably related to tumor growth and vascular development in CRC.27 Xu et al confirmed that lncRNA SNHG1 depletion inhibited the growth of CRC cells through regulating miR-154-5p.28 Therefore, it is of great importance to investigate the functional mechanism of lncRNAs for the treatment of CRC.
Increasing studies revealed that lncRNA CRNDE positively mediated cell proliferation and mobility, and repressed cell apoptosis in human cancers.29–31 However, the function of CRNDE in CRC was less studied. In this study, we confirmed that CRNDE was highly expressed in CRC tissues and cells. These results were in agreement with previous data.8 Moreover, CRNDE was reported to promote the proliferation and increase the chemotherapy of CRC cells by regulating the expression of downstream genes.13,32 Consistent with these data, our results suggested that CRNDE knockdown repressed cell proliferation, mobility, and chemotherapy against PTX, and induced apoptosis in CRC cells. From these data, we concluded that CRNDE acted as a positive regulator for the development of CRC.
It is widely accepted that lncRNAs, as a group of miRNA sponges, regulate the levels of downstream genes via binding to target genes.33 We next predicted the target genes of CRNDE using online tool starbase and found that miR-126-5p was a potential target of CRNDE. Then, this interaction was confirmed by the dual-luciferase reporter assay. Furthermore, the negative effect of CRNDE on miR-126-5p expression was observed in this research. Besides, we found that the miR-126-5p level was significantly declined in CRC tissues and cells. This was in line with previous data.18 MiR-126-5p was reported to serve as a tumor suppressor in many human cancers, such as atherosclerosis,34 cervical cancer,35 giant cell tumor,36 and non-small cell lung cancer.37 In this study, we also confirmed that miR-126-5p overexpression inhibited the growth of CRC cells and decreased drug resistance. Taken together, we guessed that CRNDE regulated CRC cell progression by modulating miR-126-5p expression. Then, this hypothesis was confirmed by our experiments, which suggested that the overexpression of CRNDE reversed the effect of miR-126-5p overexpression on CRC cell progression. Besides, it was possible that CRNDE regulated cell development by targeting other miRNAs in CRC cells. Therefore, more experiments should be performed to explore the functional mechanism of CRNDE in CRC cells.
MiRNAs are identified as a kind of regulators that modulate downstream gene expression via targeting the 3ʹUTR of target genes.38 Subsequently, online tool starbase was used to predict the targets of miR-126-5p, and then ATAD2 was found as a potential target of miR-126-5p. Then, this interaction was verified by the dual-luciferase reporter assay. Moreover, we also confirmed that miR-126-5p downregulated ATAD2 expression in CRC cells. It was reported that the ATAD2 level was dramatically upregulated in CRC.24 This was consistent with our results. Present evidence indicated that ATAD2 acted as an oncogene to promote cell growth in human cancers, including cervical cancer,39 hepatocellular carcinoma,40 gastric carcinoma,41 and papillary thyroid cancer.42 Besides, ATAD2 knockdown was reported to negatively mediate the development of CRC cells.43,44 Consistent with these data, our results suggested that the downregulation of ATAD2 suppressed cell chemotherapy, proliferation, and mobility, as well as promoted apoptosis in CRC cells. Moreover, we confirmed that miR-126-5p mediated ATAD2 expression to regulate CRC cell progression. Finally, the mechanism that CRNDE repressed miR-126-5p expression to upregulate the level of ATAD2 was confirmed in the present study.
In summary, CRNDE knockdown inhibited cell growth and PTX resistance of CRC cells through regulation of miR-126-5p/ATAD2 axis. Our findings provided a potential therapeutic target for the treatment of CRC.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
2. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254
4. Rosello S, Papaccio F, Roda D, et al. The role of chemotherapy in localized and locally advanced rectal cancer: a systematic revision. Cancer Treat Rev. 2018;63:156–171. doi:10.1016/j.ctrv.2018.01.001
5. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-38
6. Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–1933. doi:10.1111/cas.13342
7. Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol. 2014;27(10):1310–1320. doi:10.1038/modpathol.2014.33
8. Gao H, Song X, Kang T, et al. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205–216. doi:10.2147/OTT.S116178
9. Shahab SW, Matyunina LV, Mezencev R, et al. Evidence for the complexity of microRNA-mediated regulation in ovarian cancer: a systems approach. PLoS One. 2011;6(7):e22508. doi:10.1371/journal.pone.0022508
10. Zheng J, Liu X, Wang P, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–1215. doi:10.1038/mt.2016.71
11. Meng Y, Li Q, Li L, et al. The long non-coding RNA CRNDE promotes cervical cancer cell growth and metastasis. Biol Chem. 2017;399(1):93–100. doi:10.1515/hsz-2017-0199
12. Jing H, Xia H, Qian M, et al. Long noncoding RNA CRNDE promotes non-small cell lung cancer progression via sponging microRNA-338-3p. Biomed Pharmacother. 2019;110:825–833. doi:10.1016/j.biopha.2018.12.024
13. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1
14. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1–19. doi:10.1093/bib/bbs075
15. Trionfini P, Benigni A. MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol. 2017;28(6):1686–1696. doi:10.1681/ASN.2016101117
16. Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18(3):244–252. doi:10.1097/PPO.0b013e318258b78f
17. Slaby O, Svoboda M, Michalek J, et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8(1):102. doi:10.1186/1476-4598-8-102
18. Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38(14):2627–2644. doi:10.1038/s41388-018-0628-y
19. Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure–diverse function. Genes Cells. 2001;6(7):575–597. doi:10.1046/j.1365-2443.2001.00447.x
20. Ntranos A, Casaccia P. Bromodomains: translating the words of lysine acetylation into myelin injury and repair. Neurosci Lett. 2016;625:4–10. doi:10.1016/j.neulet.2015.10.015
21. Cattaneo M, Morozumi Y, Perazza D, et al. Lessons from yeast on emerging roles of the ATAD2 protein family in gene regulation and genome organization. Mol Cells. 2014;37(12):851–856. doi:10.14348/molcells.2014.0258
22. Wu S, Han M, Zhang C. Overexpression of microRNA-186 inhibits angiogenesis in retinoblastoma via the hedgehog signaling pathway by targeting ATAD2. J Cell Physiol. 2019;234(10):19059–19072.
23. Ji S, Su X, Zhang H, et al. MicroRNA-372 functions as a tumor suppressor in cell invasion, migration and epithelial-mesenchymal transition by targeting ATAD2 in renal cell carcinoma. Oncol Lett. 2019;17(2):2400–2408. doi:10.3892/ol.2018.9871
24. Hou M, Huang R, Song Y, et al. ATAD2 overexpression is associated with progression and prognosis in colorectal cancer. Jpn J Clin Oncol. 2016;46(3):222–227. doi:10.1093/jjco/hyv195
25. Chen D, Sun Q, Cheng X, et al. Genome-wide analysis of long noncoding RNA (lncRNA) expression in colorectal cancer tissues from patients with liver metastasis. Cancer Med. 2016;5(7):1629–1639. doi:10.1002/cam4.738
26. Cui C, Zhai D, Cai L, et al. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939Mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50(3):992–1008. doi:10.4143/crt.2017.226
27. Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35(3):1385–1388.
28. Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141.
29. Tang D, Zhao L, Peng C, et al. LncRNA CRNDE promotes hepatocellular carcinoma progression by upregulating SIX1 through modulating miR-337-3p. J Cell Biochem. 2019;120(9):16128–16142. doi:10.1002/jcb.28894
30. Zhu L, Yang N, Du G, et al. LncRNA CRNDE promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells via enhancing the Wnt/beta-catenin signaling pathway. J Cell Biochem. 2018.
31. Zhu L, Liu Y, Chen Q, et al. Long-noncoding RNA colorectal neoplasia differentially expressed gene as a potential target to upregulate the expression of IRX5 by miR-136-5P to promote oncogenic properties in hepatocellular carcinoma. Cell Physiol Biochem. 2018;50(6):2229–2248. doi:10.1159/000495084
32. Ding J, Li J, Wang H, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. doi:10.1038/cddis.2017.328
33. Lopez-Urrutia E, Bustamante Montes LP, Ladron De Guevara Cervantes D, et al. Crosstalk between long non-coding RNAs, Micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front Oncol. 2019;9:669. doi:10.3389/fonc.2019.00669
34. Chen Z, Pan X, Sheng Z, et al. Baicalin suppresses the proliferation and migration of Ox-LDL-VSMCs in atherosclerosis through upregulating miR-126-5p. Biol Pharm Bull. 2019;42(9):1517–1523. doi:10.1248/bpb.b19-00196
35. Wang C, Zhou B, Liu M, et al. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25(4):463–470. doi:10.3727/096504016X14685034103879
36. Wu Z, Yin H, Liu T, et al. MiR-126-5p regulates osteoclast differentiation and bone resorption in giant cell tumor through inhibition of MMP-13. Biochem Biophys Res Commun. 2014;443(3):944–949. doi:10.1016/j.bbrc.2013.12.075
37. Shi H, Bi H, Sun X, et al. Tubeimoside-1 inhibits the proliferation and metastasis by promoting miR-126-5p expression in non-small cell lung cancer cells. Oncol Lett. 2018;16(3):3126–3134. doi:10.3892/ol.2018.9051
38. Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–511. doi:10.1101/gad.1184404
39. Zheng L, Li T, Zhang Y, et al. Oncogene ATAD2 promotes cell proliferation, invasion and migration in cervical cancer. Oncol Rep. 2015;33(5):2337–2344. doi:10.3892/or.2015.3867
40. Wu G, Liu H, He H, et al. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 2014;14(1):107. doi:10.1186/1471-2407-14-107
41. Hong S, Bi M, Chen S, et al. MicroRNA-520f suppresses growth of gastric carcinoma cells by target ATPase family AAA domain-containing protein 2 (ATAD2). Neoplasma. 2016;63(6):873–879. doi:10.4149/neo_2016_606
42. Sun W, Lan X, Zhang H, et al. NEAT1_2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018;9(3):380. doi:10.1038/s41419-018-0418-z
43. Hong S, Chen S, Wang X, et al. ATAD2 silencing decreases VEGFA secretion through targeting has-miR-520a to inhibit angiogenesis in colorectal cancer. Biochem Cell Biol. 2018;96(6):761–768. doi:10.1139/bcb-2018-0081
44. Hong S, Bi M, Yan Z, et al. Silencing of ATPase family AAA domain-containing protein 2 inhibits migration and invasion of colorectal cancer cells. Neoplasma. 2016;63(6):846–855. doi:10.4149/neo_2016_603
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.