Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA Cancer Susceptibility 9 (CASC9) Up-Regulates the Expression of ERBB2 by Inhibiting miR-193a-5p in Colorectal Cancer
Authors Ding Y, Li X, Zhang Y, Zhang J
Received 14 October 2019
Accepted for publication 18 December 2019
Published 20 February 2020 Volume 2020:12 Pages 1281—1292
DOI https://doi.org/10.2147/CMAR.S234620
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
This paper has been retracted.
Yuansheng Ding, Xiaoyan Li, Yucui Zhang, Jie Zhang
The First Department of General Surgery, Linyi Central Hospital, Linyi 276400, Shandong Province, People’s Republic of China
Correspondence: Jie Zhang
The First Department of General Surgery, Linyi Central Hospital, Jiankang Road No. 17, Yishui, Linyi 276400, Shandong, People’s Republic of China
Email [email protected]
Background: Emerging studies have reported that long non-coding RNAs (lncRNAs) were crucial regulators in the progression of colorectal cancer (CRC). LncRNA susceptibility 9 (CASC9) was involved in several cancers; however, its role in CRC remains unknown.
Methods: RT-PCR was done to probe the expression of CASC9 and miR-193a-5p in CRC samples. CRC cell lines (HCT116 and SW480) were used as cell models. The biological influence of CASC9 on cancer cells was studied using CCK-8 assay, Transwell assay and TUNEL assay in vitro, and subcutaneous xenotransplanted tumor model in vivo. Interaction between CASC9 and miR-193a-5p was investigated by bioinformatics analysis, RT-PCR, and luciferase reporter assay. The expression level of the downstream gene of miR-193a-5p, erb-b2 receptor tyrosine kinase 2 (ERBB2), was tested by Western blot.
Results: CASC9 was significantly up-regulated in CRC samples, while miR-193a-5p was markedly down-regulated. Overexpression of CASC9 promoted viability, migration and invasion of CRC cells, while overexpression of miR-193a-5p had the opposite effect. CASC9 could down-regulate miR-193a-5p via sponging it, and there was a negative relevancy between CASC9 and miR-193a-5p in CRC samples. CASC9 also enhanced the expression levels of ERBB2, while this effect could be reversed by co-transfection with miR-193a-5p.
Conclusion: CASC9, an oncogenic lncRNA, was abnormally up-regulated in CRC tissues, and it could indirectly modulate the expression of ERBB2 via reducing the expression level of miR-193a-5p.
Keywords: colorectal cancer, long non coding RNA cancer susceptibility 9, CASC9, microRNA-193a-5p, erb-b2 receptor tyrosine kinase 2, ERBB2
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide. In 2018, its morbidity ranked fourth among all tumors in the world.1 Although there are many treatment strategies for CRC, the prognosis of patients who have metastasis or cannot undergo surgical resection is still unsatisfactory.2 Distant metastasis and recurrence of CRC are still the main causes of death.3,4 Accordingly, it is of great importance to further dig out the molecular mechanism in the progression of CRC for the development of new and efficient therapies for CRC.
Long non-coding RNA (LncRNA) is a class of RNA containing over 200 nucleotides, without protein-coding capacity. It has been found that lncRNA is enrolled in biological processes like chromatin modification, transcriptional interference and proto-oncogene activation, thus regulating gene expression at various levels including epigenetics, transcription or post-transcription. In recent years, many lncRNAs have been found to be involved in the progression of CRC. For instance, lncRNA XIRP2-AS1 has been found to decrease abnormally in CRC tissues, impede the proliferation and migration of CRC cells, and the progression of CRC.5 LncRNA CALIC can facilitate the metastasis of CRC cell line HCT116.6 LncRNA susceptibility 9 (CASC9) is located on chromosome 8q21.13. In ovarian cancer, high expression of CASC9 indirectly increases the expression of LIN7A by inhibiting the expression of miR-758-3p and promotes the proliferation, migration and invasion of ovarian cancer.7 However, there are few reports about the role and regulatory mechanisms of CASC9 in CRC.
MicroRNAs are a class of conservative RNAs with a length of about 18–25 nucleotides with no coding abilities. MicroRNAs are expected to become new highly sensitive markers for clinical diagnosis and disease assessment. In CRC, a variety of microRNAs have been found to be involved in the disease progression, showing anti-cancer or carcinogenic effects. For example, miR-378,8 miR-1369 inhibits the progression of CRC, while miR-183,10 miR-590-3p11 have the opposite effect. It has been proven that miR-193a-5p is a tumor suppressor in CRC.12 However, the specific regulatory mechanism of miR-193a-5p in CRC has not yet been clarified.
The gene of erb-b2 receptor tyrosine kinase 2 (ERBB2, also known as HER2) is located on chromosome 17q12-21.32 and it encodes transmembrane receptor-like protein. ERBB2 is often considered as a proto-oncogene. Over-expression of ERBB2 has been proven to significantly promote the progression of breast cancer13 and gastric cancer,14 and has become an important biomarker and therapeutic target for some patients. ERBB2 was also considered as a potential therapeutic target for CRC.15,16 Whereas, how ERBB2 is regulated during the progression of CRC remains unclear.
Through the StarBase database, we found that miR-193a-5p was one of the potential targets of CASC9, and TargetScan database showed that miR-193a-5p could target ERBB2. Based on the existing research and data, we hypothesized that CASC9 was involved in the development of CRC and could advance the progression of CRC by targeting miR-193a-5p/ERBB2 axis. This study aims to figure out the roles of CASC9, miR-193a-5p and ERBB2 in CRC and their mutual regulation mechanisms, so as to further expound the molecular basis of the occurrence and progression of CRC and to make a contribution to the exploration of new therapeutic targets.
Materials and Methods
Clinical Samples
Human samples collection and the subsequent experiments were performed according to the Declaration of Helsinki. All patients provided written informed consent. CRC tissue samples and adjacent normal tissues were collected from 70 patients with CRC who were treated in our hospital from January 2017 to April 2018. The samples had complete clinical and pathological records. All patients signed the informed consent, and the project was under the approval of the Ethics Committee of Linyi Central Hospital.
Cell Culture
CRC cell lines HCT116, SW480 and normal human colon epithelial cell NCM460 were purchased from the Cell Center of the Chinese Academy of Medical Sciences, Shanghai, China. The above cells were cultured in DMEM medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Grand Island, NY, USA) and 1% penicillin/streptomycin, and cultured at 37°C in 5% CO2. Cells in logarithmic growth phase were used for the subsequent experiments.
Cell Transfection
HCT116 cells and SW480 cells in the logarithmic growth phase were inoculated into 6-well plates with cell density of 5×105/well. When the cell growth reached 50–60% fusion, pcDNA3.1-CASC9 plasmid, pcDNA3.1 plasmid or miR-193a-5p mimics, mimics-NC were transfected into HCT116 cells and CASC9-shRNA, shRNA-NC or miR-193a-5p inhibitors or inhibitors-NC were transfected into SW480 cells according to the instructions of Lipofectamine2000 (Invitrogen; Thermo Fisher Scientific, Inc., CA, USA). Forty-eight hours after transfection, the cells were harvested for the subsequent experiments.
qRT-PCR
TRIzol (Invitrogen, Carlsbad, CA, USA) was employed to extract the total RNA from tissues and cells. In compliance with the supplier’s instructions, total RNA was reversely transcribed into cDNA with PrimeScript RT Reagent Kit (Invitrogen, Shanghai, China). Bio-Rad CFX96 and SYBR Green Premix Ex Taq II (Takara, Dalian, China) were adopted for RT-PCR, which was carried out according to the manufacturer’s regulations. GAPDH and U6 were used as reference genes, and 2(-ΔΔCt) method was used for statistics. The specific primer sequence information is shown in Table 1.
 |
Table 1 The Sequence of PCR Primers Used |
CCK-8 Assay
HCT116 and SW480 cells in the logarithmic growth phase were harvested. After adjusting the cell density to 2×104/mL, the cells were inoculated into a 96-well plate with 100 μL cell suspension per well. After that, the 96-well plate was placed in an incubator for further culture. After 24 hrs, CCK-8 kit (Dojindo, Kumamoto, Japan) was added into each well and cultured in an incubator for another 1 hr. After that, 96-well plates were placed in a microplate reader to determine the OD value of each well at 450 nm wavelength. The OD values of cells were measured at 24, 48, 72 and 96 h, respectively, and the proliferation curve was plotted.
Transwell Assay
5×104 transfected cells were seeded in the upper chamber of the Transwell system (Corning, Shanghai, China) with FBS-free medium and medium with 10% FBS was added in the lower chamber. After culture at 37°C for 24 hrs, the cells failing to pass through the membrane were discarded from the upper chamber. Then, the membrane was fixed with 4% paraformaldehyde for 10 mins and stained with 0.5% crystal violet. Following washing with running water gently, the cells were observed under a microscope. Matrigel was used in the invasion experiment, but not in the migration experiment.
TUNEL Assay
Cells in each group were cultured on gelatin-coated coverslips. Cells were washed with PBS and then fixed in 4% paraformaldehyde. Next, the samples were incubated at 37°C for 60 mins with 50 μL TUNEL detection kit (Beyotime, Hangzhou, China) according to the manufacturer’s protocols and then washed with PBS for three times. The liquid seal was quenched with anti-fluorescence and observed under a fluorescence microscope: the excitation light wave was 450–500 nm and the emission light was 515–565 nm (green fluorescence). Five visual fields were randomly selected and observed for each sample and the apoptotic rate was calculated: apoptotic rate = apoptotic cells/total cells × 100%.
Western Blot
RIPA lysis buffer was utilized to extract total proteins from different groups of cells. Then, protein concentration was determined by BCA method. Total proteins were separated by SDS-PAGE electrophoresis and transferred to PVDF membranes. Then, the membranes were blocked with 5% skim milk for 1 hr, and then the membrane was incubated with primary antibody (ab16901, 1:1000, abcam) at 4°C for overnight. After that, TBST was used to rinse the membrane, and Horseradish peroxidase labelled secondary antibody (ab205719, 1:2000, abcam) was added and incubated at 37°C for 1 hr. At last, the band was developed with chemiluminescence using hypersensitive ECL (Guangzhou Xiangbo Biotechnology Co., Ltd.).
Luciferase Reporter Assay
Luciferase reporter assay was used to validate the targeting relationships between miR-193a-5p and CASC9 or 3ʹ-UTR of ERBB2. The wild type (WT) CASC9 sequence or the WT 3ʹ-UTR fragment from ERBB2 mRNA including the predicted binding site of miR-193a-5p were amplified and inserted into the pmirGLO dual-luciferase microRNA target expression vector (Promega, Madison, WI, USA) to construct the report vector pmirGLO-CASC9-WT or pmirGLO-ERBB2-WT. The presumed binding sites of miR-193a-5p in CASC9 or ERBB2 3ʹ-UTR were mutated by GeneArt Site-Directed Mutagenesis PLUS System (cat. No. A14604; Thermo Fisher Science, Inc.). Mutant (MUT) CASC9 sequence or ERBB2 3′-UTR sequence were inserted into the pmirGLO vector to construct report vector pmirGLO-CASC9-MUT or pmirGLO-ERBB2-MUT. The corresponding reporter vectors and miR-193a-5p or NC mimics were co-transfected into SW480 and HCT116 cells and incubated for 48 hrs. Luciferase activity was then measured via Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Tumorigenesis in Nude Mice
The protocols of the animal experiments were approved by the Animal Care and Use Committee of Linyi Central Hospital. The experiment was conducted in accordance with UKCCCR Guidelines for the welfare of animals in experimental neoplasia. 2×107 HCT116 cells in lentivirus-CASC9 group, lentivirus-negative control group and empty control group were subcutaneously injected into the right back of female BALB/c nude mice (n=5). All the nude mice were successfully implanted and the tumor formation rate reached 100%. Tumor volume was observed and measured once a week after subcutaneous injection. Tumor-carrying mice were executed 49 days later. Tumor volume (mm3) and weight (g) were measured after removal of tumors. The volume of tumors was calculated under the following formula: volume = length × width2 × 0.5.
Statistical Analysis
Statistical software SPSS 17.0 (SPSS Inc., Chicago, IL, USA) was applied for analyzing all the experimental data in this study. The data were expressed as mean ±standard deviation (x±s). The differences between two or more groups were analyzed by Student’s t-test. Chi-square test was conducted to compare differences of enumeration data in different groups. P < 0.05 expressed statistically significant.
Results
Abnormal Expression of CASC9 and miR-193a-5p in CRC Tissues and Cell Lines
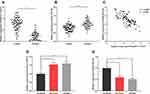
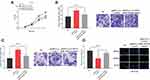
To preliminarily explore the role of CASC9 and miR-193a-5p, we compared the expression levels of CASC9 and miR-193a-5p in CRC tissues with those in adjacent normal tissues by qRT-PCR. As shown, the expression of CASC9 was significantly increased in CRC tissues, while the expression of miR-193a-5p was down-regulated (Figure 1A and B). Further analysis found that the expression levels of CASC9 and miR-193a-5p were negatively correlated in CRC tissues (Figure 1C). We also measured the expression of CASC9 and miR-193a-5p in CRC cell lines HCT116 and SW480. We found that, compared with NCM460 cells, CASC9 was significantly up-regulated and miR-193a-5p was significantly down-regulated in CRC cell lines (Figure 1D and E). These results indicated that in CRC, CASC9 probably functioned as an oncogenic lncRNA, while miR-193a-5p might be a tumor suppressor. Additionally, we also analyzed the correlation between the expression of CASC9 and the pathological characteristics of the patients, and found that the expression of CASC9 was not related to the age and sex of patients, but related to the TNM stage of CRC (Table 2). This further suggested that CASC9 was associated with the progression of CRC.
 |
Table 2 The Relationship Between Characteristic CASC9 Expression in Patients with CRC |
CASC9 and miR-193a-5p Affected Proliferation, Migration, Invasion and Apoptosis of CRC Cells
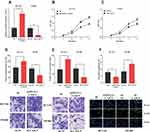
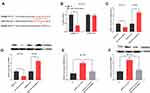
To explore the function of CASC9, we successfully constructed the CASC9 over-expression and knockdown cell lines with HCT116 and SW480 cells, respectively (Figure 2A). Through CCK-8 and Transwell experiments, we demonstrated that the proliferation, migration and invasion of HCT116 cells with high expression of CASC9 were significantly enhanced. In the TUNEL assay, the apoptosis of HCT116 cells with high expression of CASC9 was reduced. In SW480 cells, knockdown of CASC9 inhibited proliferation, migration and invasion of SW480 cells. In the TUNEL experiment, we found that CASC9 knockdown enhanced the apoptosis of SW480 cells (Figure 2B–F). Collectively, CASC9 played a significant role in regulating proliferation, migration and apoptosis of CRC cells.
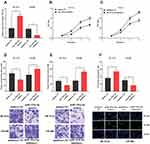
To explore the function of miR-193a-5p, we successfully constructed models of over-expression and low expression of miR-193a-5p by transfecting miR-193a-5p mimics and miR-193a-5p inhibitors into HCT116 and SW480 cells, respectively (Figure 3A). Similarly, CCK-8, Transwell and TUNEL experiments were used to investigate the proliferation, metastasis and apoptosis. We found that the transfection of miR-193a-5p mimics significantly suppressed the proliferation, migration and invasion of HCT116 cells, and promoted the apoptosis of HCT116 cells. Conversely, the transfection of miR-193a-5p inhibitors could promote the proliferation, migration and invasion of SW480 cells and inhibit their apoptosis (Figure 3B–F). It could be concluded that miR-193a-5p exerted a tumor-suppressive role in the malignant phenotypes of CRC cells.
CASC9 Targeted miR-193a-5p
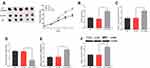
Previous studies have shown that lncRNA could function as a competitive endogenous RNA (ceRNA) in the regulation of the function of miRNAs. As mentioned above, CASC9 and miR-193a-5p were negatively correlated in CRC tissues (Figure 1C). Meanwhile, CASC9 and miR-193a-5p had opposite effects on cell proliferation, apoptosis, migration and invasion (Figures 2 and 3). Therefore, we were curious about the regulatory relationship between CASC9 and miR-193a-5p. Through DIANA tools, we found a potential binding site between CASC9 and miR-193a-5p (Figure 4A). The results of dual-luciferase reporter assay showed that the activity of luciferase of wild type CASC9 sequence was significantly decreased by miR-193a-5p, while the mutant CASC9 sequence was not affected (Figure 4B and C). This confirmed the binding relationship between CASC9 and miR-193a-5p. Additionally, in CRC cells, the expression of miR-193a-5p was decreased when CASC9 was overexpressed, but increased when CASC9 was knocked down (Figure 4D). Therefore, we concluded that CASC9 could target and down-regulate miR-193a-5p in CRC. To further validate the interaction between CASC9 and miR-193a-5p in CRC progression, we transfected miR-193a-5p mimics into HCCT116 cells with CASC9 overexpression. It was found that the function of CASC9 in promoting the proliferation of HCT116 cells was significantly reversed by miR-193a-5p mimics. In the Transwell experiment, miR-193a-5p also reversed the role of CASC9 in promoting CRC cell migration and invasion (Figure 5A–C). In addition, the inhibitory effect of CASC9 on apoptosis of HCT116 cells was also attenuated by miR-193a-5p mimics (Figure 5D).
CASC9 Indirectly Regulated ERBB2 Through miR-193a-5p
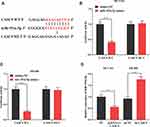
To further explore the mechanism of the action of miR-193a-5p, we predicted the downstream target of miR-193a-5p by TargetScan and found that there was a binding site between miR-193a-5p and ERBB2 (Figure 6A). Dual-luciferase reporter assay validated the binding relationship between miR-193a-5p and ERBB2 (Figure 6B). In addition, we found that miR-193a-5p mimics could significantly inhibit the expression of ERBB2 mRNA and protein, while miR-193a-5p inhibitors showed the opposite effect (Figure 6C and D). What is more, in HCT116 cells with CASC9 over-expression, we observed a significant increase in ERBB2 mRNA and protein, whereas miR-193a-5p mimics inhibited them (Figure 6E and F). Collectively, these results suggested that ERBB2 could be inhibited by miR-193a-5p and indirectly regulated by CASC9.
Over-Expression of CASC9 Promoted the Growth of Transplanted Tumors in Nude Mice
To further explore the effect of CASC9 on CRC progression, we constructed a xenotransplantation model of CRC by injecting CASC9-overexpressed HCT116 cells subcutaneously into BALB/c nude mice. The results showed that, compared with the control groups, the volume and weight of the tumors in the CASC9 overexpression group increased significantly (Figure 7A and B). qRT-PCR and Western blot indicated that, in the tumor tissues, CASC9 expressions were remarkably higher in the CASC9 over-expression group than in the control groups (Figure 7C), and the expression of miR-193a-5p was markedly reduced (Figure 7D), and the expression levels of ERBB2 mRNA and protein were in a significant increase (Figure 7E and F). So far, we confirmed that CASC9 could promote the progression of CRC by targeting the miR-193a-5p/ERBB2 axis.
Discussion
LncRNA plays an important regulatory role in many biological processes of cancer cells, including tumorigenesis, proliferation, apoptosis, metastasis, chemoresistance, radioresistance, stemness, etc.17 For example, the high expression of lncRNA SNHG7 in gastric cancer promotes the proliferation of cancer cells while inhibiting their apoptosis.18 In pancreatic cancer, lncRNA HOTTIP enhances the stem cell characteristics of cancer cells and facilitates cancer progression.19 In nasopharyngeal carcinoma, down-regulation of lncRNA ANRIL was found to increase the sensitivity of tumor cells to radiotherapy.20 Herein we demonstrated that the expression of CASC9 was abnormally increased in CRC tissues and cell lines, and was correlated with the pathological indexes of CRC. CCK-8, Transwell and TUNEL experiments showed that up-regulation of CASC9 enhanced the proliferation, migration and invasion, but inhibited the apoptosis of CRC cells, while down-regulation of CASC9 resulted in the opposite effects. In vivo experiments also indicated that CASC9 significantly promoted the growth of xenotransplantated tumors. These data suggested that CASC9 played an oncogenic role in CRC.
Recently, studies have confirmed that lncRNAs, as ceRNA of some microRNAs, formed a huge and complex regulatory network. For example, in tongue squamous cell carcinoma, lncRNA MALAT1 can down-regulate SLAIN2 and reduce the invasion and metastasis of cancer cells through sponging miR-106b-5p;21 in cervical cancer cells, lncRNA MALAT1 acts as ceRNA of miR-124 to promote cancer progression.22 In ovarian cancer, CASC9 can indirectly regulate LIN7A and promote the progression of cancer by targeting miR-758-3p.7 In this study, miR-193a-5p identified as a downstream target of CASC9. Our analysis demonstrated that CASC9 could act as a ceRNA of miR-193a-5p and possibly exerted its cancer-promoting function through repressing miR-193a-5p. In addition to regulate ceRNA network, CASC9 was also found to interact directly with a variety of proteins. For example, in esophageal squamous cell carcinoma, the up-regulation of CASC9 reduces the expression level of PDCD4 by recruiting EZH2, thus promoting tumor growth.23 In the future, other downstream miRNAs of CASC9 needs to be identified, and it is quite interesting to investigate whether CASC9 plays its role via other mechanisms except “ceRNA” in CRC. In addition to lncRNA, miRNAs are another kind of non-coding RNAs with remarkable tumor regulation function. By binding to the 3ʹUTR of the coding gene’s mRNA, miRNAs can control the expression of target proteins at post-transcriptional and translation stages.24,25 miR-193a-5p was usually considered as an anti-cancer factor. Its anti-cancer effects in hepatocellular carcinoma,26 glioblastoma,27 breast cancer28 and other cancers have been confirmed. In our study, miR-193a-5p has been shown to inhibit the malignant phenotypes of CRC cells, thereby inhibiting the progression of CRC. Other studies have confirmed that miR-193a-5p could hinder the migration of CRC cell HT-29 by inhibiting metastasis-related pathways, which was consistent with our demonstrations.12
ErbB family members activate the receptor cytoplasmic tyrosine kinase domain by forming homologous or heterodimer. Abnormal activation of ErbB family proteins can recruit a variety of downstream signal proteins, and ultimately activate Ras/Raf/MAPK, ERK1/ERK2 signaling pathways, thereby promoting cancer cell proliferation, migration and invasion. ERBB2 is the first choice for the dimerization of other receptors.29 In breast cancer, ERBB2 is negatively regulated by miR-155, promoting the malignant transformation of breast epithelial cells;13 in non-small-cell lung cancer, miR-331-3p inhibits epithelial–mesenchymal transition partly by targeting ERBB2.30 In this study, ERBB2 was identified as a target gene of miR-193a-5p and could be negatively regulated by it. Previous studies and our findings imply that the dysregulation of miRNAs could be an important reason for ERBB2 activation in cancers.
In conclusion, our findings demonstrated the abnormal high expression of CASC9 in CRC and its clinical significance. It was also revealed that CASC9 promoted the malignant phenotypes of CRC cells through adsorbing miR-193a-5p and up-regulating ERBB2. Our study further revealed the molecular regulatory mechanism in the development of CRC, and could provide new potential therapeutic targets for this disease.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
Our study was approved by the Ethics Committee of Linyi Central Hospital and was conducted in accordance with the Declaration of Helsinki. The protocols of the animal experiments were approved by the Animal Care and Use Committee of Linyi Central Hospital.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.v69.1
2. Byrne M, Saif MW. Selecting treatment options in refractory metastatic colorectal cancer. Onco Targets Ther. 2019;12:2271–2278. doi:10.2147/OTT.S194605
3. Gao Q, Tsoi KK, Hirai HW, et al. Serrated polyps and the risk of synchronous colorectal advanced neoplasia: a systematic review and meta-analysis. Am J Gastroenterol. 2015;110(4):501–509. doi:10.1038/ajg.2015.49
4. Elwood PC, Morgan G, Pickering JE, et al. Aspirin in the treatment of cancer: reductions in metastatic spread and in mortality: a systematic review and meta-analyses of published studies. PLoS One. 2016;11(4):e0152402. doi:10.1371/journal.pone.0152402
5. Zhou F, Shen F, Zheng Z, Ruan J. The LncRNA XIRP2-AS1 predicts favorable prognosis in colon cancer. Onco Targets Ther. 2019;12:5767–5778. doi:10.2147/OTT.S215419
6. Kawasaki Y, Miyamoto M, Oda T, et al. The novel lncRNA CALIC upregulates AXL to promote colon cancer metastasis. EMBO Rep. 2019;20(8):e47052. doi:10.15252/embr.201847052
7. Hu X, Li Y, Kong D, Hu L, Liu D, Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234(7):10800–10808. doi:10.1002/jcp.v234.7
8. Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22:12. doi:10.1186/s11658-017-0041-5
9. Yuan Q, Cao G, Li J, Zhang Y, Yang W. MicroRNA-136 inhibits colon cancer cell proliferation and invasion through targeting liver receptor homolog-1/Wnt signaling. Gene. 2017;628:48–55. doi:10.1016/j.gene.2017.07.031
10. Bi DP, Yin CH, Zhang XY, Yang NN, Xu JY. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep. 2016;35(5):2873–2879. doi:10.3892/or.2016.4631
11. Feng ZY, Xu XH, Cen DZ, Luo CY, Wu SB. miR-590-3p promotes colon cancer cell proliferation via Wnt/β-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur Rev Med Pharmacol Sci. 2017;21(21):4844–4852.
12. Shirafkan N, Shomali N, Kazemi T, et al. microRNA-193a-5p inhibits migration of human HT-29 colon cancer cells via suppression of metastasis pathway. J Cell Biochem. 2018. doi:10.1002/jcb.28164
13. He XH, Zhu W, Yuan P, et al. miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene. 2016;35(46):6015–6025. doi:10.1038/onc.2016.132
14. Shan X, Wen W, Zhu D, et al. miR 1296-5p inhibits the migration and invasion of gastric cancer cells by repressing ERBB2 expression. PLoS One. 2017;12(1):e0170298. doi:10.1371/journal.pone.0170298
15. Ma EL, Choi YJ, Choi J, Pothoulakis C, Rhee SH, Im E. The anticancer effect of probiotic Bacillus polyfermenticus on human colon cancer cells is mediated through ErbB2 and ErbB3 inhibition. Int J Cancer. 2010;127(4):780–790. doi:10.1002/ijc.25011
16. Huang Y, Sun Y, Wang WW, Zhang L. Boeravinone B a natural rotenoid exerts anticancer activity via inducing internalization and degradation of inactivated EGFR and ErbB2 in human colon cancer cells. Am J Transl Res. 2018;10(12):4183–4192.
17. Jiang P, Han X, Zheng Y, Sui J, Bi W. Long non-coding RNA NKILA serves as a biomarker in the early diagnosis and prognosis of patients with colorectal cancer. Oncol Lett. 2019;18(2):2109–2117. doi:10.3892/ol.2019.10524
18. Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–4622.
19. Fu Z, Chen C, Zhou Q, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi:10.1016/j.canlet.2017.09.019
20. Hu X, Jiang H, Jiang X. Downregulation of lncRNA ANRIL inhibits proliferation, induces apoptosis, and enhances radiosensitivity in nasopharyngeal carcinoma cells through regulating miR-125a. Cancer Biol Ther. 2017;18(5):331–338. doi:10.1080/15384047.2017.1310348
21. Zhuang M, Zhao S, Jiang Z, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi:10.1016/j.ebiom.2018.12.049
22. Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016;37(1):633–640. doi:10.1007/s13277-015-3732-4
23. Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16(1):150. doi:10.1186/s12943-017-0715-7
24. Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461(7263):546–549. doi:10.1038/nature08349
25. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi:10.1093/nar/gkj112
26. Li P, Xiao Z, Luo J, Zhang Y, Lin L. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23(4):2475–2488. doi:10.1111/jcmm.2019.23.issue-4
27. Jin L, Li H, Wang J, et al. MicroRNA-193a-5p exerts a tumor suppressor role in glioblastoma via modulating NOVA1. J Cell Biochem. 2019;120(4):6188–6197. doi:10.1002/jcb.v120.4
28. Xie F, Hosany S, Zhong S, et al. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS One. 2017;12(10):e0185565. doi:10.1371/journal.pone.0185565
29. Subramanian J, Katta A, Masood A, Vudem DR, Kancha RK. Emergence of ERBB2 mutation as a biomarker and an actionable target in solid cancers. Oncologist. 2019;24:12. pii: theoncologist.2018–0845. doi:10.1634/theoncologist.2018-0845
30. Li X, Zhu J, Liu Y, Duan C, Chang R, Zhang C. MicroRNA-331-3p inhibits epithelial-mesenchymal transition by targeting ErbB2 and VAV2 through the Rac1/PAK1/β-catenin axis in non-small-cell lung cancer. Cancer Sci. 2019;110(6):1883–1896. doi:10.1111/cas.14014
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.