Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA AGAP2-AS1/miR-628-5p/PTN Axis Modulates Proliferation, Migration, Invasion, and Apoptosis of Glioma Cells
Authors Yan Y, Wang Y, Liu Y, Chen T, Zhu Y, Li H, Kong F
Received 22 February 2020
Accepted for publication 13 June 2020
Published 20 July 2020 Volume 2020:12 Pages 6059—6068
DOI https://doi.org/10.2147/CMAR.S250890
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
This paper has been retracted.
Yang Yan, 1,* Yiping Wang, 1,* Yuxia Liu, 2 Tao Chen, 3 Yaoli Zhu, 4 Huiqing Li, 5 Fangen Kong 1
1Department of Neurosurgery, The Fifth Affiliated Hospital Sun Yat-Sen University, Zhuhai, Guangdong, People’s Republic of China; 2Department of Gastrointestinal Surgery, The Fifth Affiliated Hospital Sun Yat-Sen University, Zhuhai, Guangdong, People’s Republic of China; 3Department of Spine Surgery, The Fifth Affiliated Hospital Sun Yat-Sen University, Zhuhai, Guangdong, People’s Republic of China; 4Department of Critical Care Medicine, The Fifth Affiliated Hospital Sun Yat-Sen University, Zhuhai, Guangdong, People’s Republic of China; 5Department of Neurology, The Fifth Affiliated Hospital Sun Yat-Sen University, Zhuhai, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fangen Kong Email [email protected]
Purpose: Long non-coding RNAs (lncRNAs) have been reported to be involved in a variety of cancers, including glioma. However, the exact role and underlying mechanism of lncRNA AGAP2 antisense RNA 1 (AGAP2-AS1) in glioma have not yet been fully elucidated.
Methods: The expression levels of AGAP2-AS1, microRNA-628-5p (miR-628-5p) and pleiotrophin (PTN) were measured by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation, apoptosis, migration and invasion were detected by Cell Counting Kit-8 (CCK-8) assay, flow cytometry, transwell assay, respectively. Western blot assay was used to detect the protein level of PTN. The interaction between miR-628-5p and AGAP2-AS1 or PTN was predicted by bioinformatics software and confirmed by the dual-luciferase reporter and RNA Immunoprecipitation (RIP) assays. Murine xenograft model was established to confirm the role of AGAP2-AS1 in glioma progression in vivo.
Results: AGAP2-AS1 expression was upregulated in glioma tissues and cells. Knockdown of AGAP2-AS1 inhibited the proliferation, migration and invasion, but facilitated apoptosis in glioma cells. Moreover, AGAP2-AS1 could directly bind to miR-628-5p and its overexpression reversed the anti-tumor effect of miR-628-5p restoration on the progression of glioma cells. In addition, miR-628-5p directly targeted PTN and its inhibition abolished the inhibitory effect of PTN knockdown on the progression of glioma cells. Furthermore, AGAP2-AS1 functioned as a competing endogenous RNA (ceRNA) by sponging miR-628-5p to modulate PTN expression. Besides, AGAP2-AS1 depletion reduced tumor growth by upregulating miR-628-5p and downregulating PTN.
Conclusion: AGAP2-AS1 knockdown suppressed cell proliferation, migration and invasion but promoted cell apoptosis in glioma cells by regulating miR-628-5p/PTN axis, providing novel avenues for treatment of glioma.
Keywords: glioma, AGAP2-AS1, miR-628-5p, PTN, cell progression
Introduction
Glioma, a kind of malignant brain tumor, is one of the most deadly cancers in adults.1 In spite of significant advance has been made in the diagnosis and treatment of glioma, the prognosis is still dismal and the median survival time for high-grade glioma patients is only 10–15 months.2–4 Hence, elucidating the pathogenic mechanisms at the molecular level and searching effective therapeutic strategies are critical for the treatment of glioma.
Long non-coding RNAs (lncRNAs), more than 200 nucleotides, play essential roles in tumorigenesis in a variety of cancers and without protein-coding ability.5–7 It has been confirmed that the aberrant expression of lncRNAs is tightly associated with diverse biological processes, such as cell differentiation, cell growth, apoptosis, cell cycle, drug resistance, metastasis, and epithelial-mesenchymal transition.8,9 LncRNAs are considered to be critical regulators of tumorigenesis and cancer progression, and more and more lncRNAs have been identified as oncogenes or tumor suppressors.10 LncRNA AGAP2 antisense RNA 1 (AGAP2-AS1), an antisense lncRNA located at 12q14.1, has been identified as an oncogene in multiple cancers, such as breast cancer,11 non-small cell lung cancer,12 and gastric cancer.13 Additionally, Zheng et al pointed out that AGAP2-AS1 abundance was enhanced in glioma tissues and cells.14 However, the involvement of AGAP2-AS1 in glioma pathogenesis remains to be explored.
Recent reports have proven that lncRNAs serve as competing endogenous RNA (ceRNA), which interact with microRNAs (miRNAs) and modulate the expression of miRNA target genes.15 MiR-628-5p is found to be a potential biomarker in several diseases, such as ovarian cancer,16 prostate cancer,17 and acute myeloid leukemia.18 Additionally, miR-628-5p has been reported to be expressed at a low level in the glioma tissues and cell lines.19 However, the interaction between AGAP2-AS1 and miR-628-5p has not been reported. Pleiotrophin (PTN), located on chromosome 7, is expressed in the brain during embryogenesis and commonly upregulated in many cancers, including glioma.20,21 Bioinformatics analysis exhibits the potential binding sites between miR-628-5p and AGAP2-AS1 or PTN. Thus, we supposed that AGAP2-AS1 might regulate the progression of glioma through acting as a sponge of miR-628-5p to regulate PTN expression. In the current study, the abundance of AGAP2-AS1 was measured in glioma tissues and cells. Moreover, the biological functions of AGAP2-AS1 in glioma cell growth, apoptosis, migration, and invasion were further investigated. Furthermore, we explored the ceRNA regulatory network of AGAP2-AS1/miR-628-5p/PTN in glioma cells. Collectively, this research showed a new ceRNA regulatory network in glioma.
Methods
Patients’ Specimens
Nontumorous brain tissues (NBTs) and glioma tissues were provided by The Fifth Affiliated Hospital Sun Yat-Sen University in this study. NBTs were collected from 14 patients who underwent a partial brain resection because of traumatic brain injury or intracerebral hemorrhage. Glioma clinical tissues were collected from 55 patients with glioma requiring surgical resection. In these tissues, 30 cases were low-grade glioma tissues (LGG; grade II) and 25 cases were high-grade glioma tissues (HGG; grade III and IV). These participants did not receive chemotherapy, radiotherapy or others’ therapy before surgery. This study was approved by the Research Ethics Committee of The Fifth Affiliated Hospital Sun Yat-Sen University. All participants signed the informed consent. Fresh samples were promptly frozen in liquid nitrogen and then stored at −80°C before use.
Cell Culture and Transfection
Two glioma cell lines (U251 and LN229) were purchased from COBIOER (Nanjing, China), and two other glioma cell lines (A172 and SHG44) were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Normal human astrocytes (NHAs) were brought from Lonza (Basel, Switzerland). These cells were grown in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100 μg/mL) (Gibco, Carlsbad, CA, USA) in a humidified atmosphere with 5% CO2 at 37°C.
The small interfering RNA against-AGAP2-AS1 or PTN (si-AGAP2-AS1 or si-PTN) and their control (si-control), miR-628-5p mimic or inhibitor (miR-628-5p or miR-628-5p inhibitor) and their control (miR-control or inhibitor-control), and AGAP2-AS1 overexpression vector (AGAP2-AS1) were obtained from GenePharma (Shanghai, China). Lentiviral small hairpin RNA targeting AGAP2-AS1 (sh-AGAP2-AS1) and its control (sh-control) were provided by Ribo Bio Corporation (Guangzhou, China). SHG44 and U251 cells were transfected with oligonucleotides or vectors using the Lipofectamine 3000 reagent (Invitrogen).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from tissue and cell lines was extracted using TRIzol reagent (Invitrogen). The first strand of complementary DNA (cDNA) for AGAP2-AS1 and PTN or miR-628-5p was synthesized with TaqMan Reverse Transcription Kit or TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), respectively. Real-time PCR was performed on a 7900HT Fast RT-PCR instrument using SYBR Green PCR Master Mix (Applied Biosystems) and following the primers: AGAP2-AS1 (Forward, 5ʹ-TACCTTGACCTTGCTGCTCTC-3ʹ; Reverse, 5ʹ-TGTCCCTTAATGACCCCATCC-3ʹ), miR-628-5p (Forward, 5ʹ-GCTGACATATTTACTAGAG-3ʹ; Reverse, 5ʹ-GAACATGTCTGCGTATCTC-3ʹ) PTN (Forward, 5ʹ-AGAAGCAATTTGGCGCGGA-3ʹ; Reverse, 5ʹ-TGCACCACCAACTGCTTAGC-3), GAPDH (Forward, 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ; Reverse, 5ʹ- ATCCGTTGACTCCGACCTTCAC-3ʹ), U6 (Forward, 5ʹ-CTCGCTTCGGCAGCACAT’; Reverse, 5ʹ-CGCAAATTCGTGAAGCGTTCCA3ʹ). Analysis of relative AGAP2-AS1, PTN or miR-628-5p expression was evaluated with 2−ΔΔCt method with normalization to GAPDH or U6, respectively. All experiments are performed three times.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) was used to determine cell proliferation capacity. In brief, SHG44 and U251 cells growing at an exponential rate were seeded in 96-well plates (4000 cells/well). CCK-8 (10 μL) solution was added to the culture medium at 0 h, 24 h, 48 h and 72 h after transfection, then incubated for an additional 3 h. After that, the absorbance was evaluated at 450 nm under a microplate reader (Bio-Rad, Hercules, CA, USA). All experiments are performed three times.
Cell Apoptosis Assay
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit double staining kit (BD Biosciences, Franklin Lakes, NJ, USA) was utilized to detect cell apoptosis. In brief, SHG44 and U251 cells growing at an exponential rate were seeded into 6 cm dishes and transfected with si-control, si-AGAP2-AS1, miR-control, miR-628-5p, miR-628-5p + AGAP2-AS1, si-PTN, or si-PTN + miR-628-5p inhibitor. After transfection for 48 h, SHG44 and U251cells were collected, washed, and re-suspended in binding buffer (500 µL), followed by staining with Annexin V-FITC and PI for 15 min. Finally, cell apoptosis was analyzed by a flow cytometer (BD Biosciences). Non-apoptotic cells were defined as Annexin V-FITC−/PI−, early apoptotic cells were defined as Annexin V-FITC+/PI−, late apoptotic/necrotic cells were defined as Annexin V-FITC+/PI+, and dead cells were defined as Annexin V-FITC−/PI+. All experiments are performed three times.
Transwell Assay
For cell invasion assay, the transfected SHG44 and U251 cells were suspended in serum-free DMEM (100 μL) and placed in the upper chamber of transwell chamber (8 μm pore size, Corning Inc., Corning, NY, USA) containing a Matrigel-coated (BD, San Jose, CA, USA), while cell migration assay did not coat with Matrigel. DMEM with 10% FBS (500 μL) was added to the lower chamber. Following incubation for 48 h, non-migrated and non-invaded cells were scrubbed by a cotton swab; migrated and invaded cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet, and counted by an inverted microscope (Olympus, Tokyo, Japan). All experiments are performed three times.
Dual-Luciferase Reporter Assay
The wild-type (WT) or mutant (MUT) of AGAP2-AS1 or 3ʹ-UTR of PTN containing the putative binding sites of miR-628-5p was synthesized and inserted into the pGL3-control luciferase reporter vectors (Promega, Madison, WI, USA), namely wild-type plasmids (WT-AGAP2-AS1, 3ʹUTR-WT-PTN) or mutant-type plasmids (MUT-AGAP2-AS1, 3ʹUTR-MUT-PTN). WT or MUT plasmid transiently co-transfected with miR-628-5p mimic or miR-control into SHG44 and U251 cells for 48 h. Finally, luciferase activity was examined using the Dual-Luciferase Assay System (Promega) and normalized to Renilla luciferase activity. All experiments are performed three times.
RNA Immunoprecipitation (RIP) Assay
EZ-Magna RIP Kit (Millipore) was applied to conduct RIP assay. In brief, SHG44 and U251 cells were lysed by a complete RIP lysis buffer. After that, whole-cell extract (100 µL) was incubated by RIP buffer containing magnetic beads coated with human anti-argonaute2 (Ago2) or anti-IgG antibody (as control). Next, samples were digested using the proteinase K. Lastly, immunoprecipitated RNA was isolated, and the expression levels of AGAP2-AS1 and miR-628-5p were examined by qRT-PCR.
Western Blot Assay
Total protein from tissues and cells (SHG44 and U251) was extracted using RIPA lysis buffer supplemented with protease (Beyotime) on ice. The protein concentration of the lysates was assessed by a bicinchoninic acid protein assay kit (Tanon, Shanghai, China). Protein from each sample (about 40 μg) was subjected to 12% SDS-PAGE (80 V for 30 min and 120 V for 40 min). Subsequently, the gels containing the goal protein were transferred onto a polyvinylidene fluoride (PVDF) membrane (0.2 μm, Beyotime). After that, the membranes containing the goal protein were blocked using 5% non-fat milk in Tris-Buffered Saline Tween-20 (TBST), followed by incubation overnight at 4°C with the specific antibodies against PTN (1:2000, ab223675, Abcam, Cambridge, MA, USA) and GAPDH (1:2000, ab37168, Abcam). After washing with TBST, secondary antibody was added to the blots for 2 h. Finally, the blots were detected with enhanced chemiluminescence reagent (Pierce, Rockford, IL, USA). The protein level of PTN was normalized by GAPDH. The intensity of the bands was analyzed by ImageJ software. All experiments are performed three times.
Tumor Xenograft Model
The animal experiments were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health” and approved by the Animal Experimental Ethics Committee of The Fifth Affiliated Hospital Sun Yat-Sen University. BALB/c nude mice (five-week-old, male) were provided by Shanghai Experimental Animal Center (Shanghai, China). The mice were randomly grouped on the basis of body weight on the day of tumor cell inoculation using the stratified randomization method (n=7/group). U251 cells transfected with sh-NC or sh-AGAP2-AS1 were subcutaneously injected into the dorsal flanks of nude mice. We examined the xenograft tumor volume every week and calculated by the following formula: length × width2/2. After 5 weeks, mice were sacrificed, and the tumors were weighed and collected for further analysis.
Statistical Analysis
All experiments were repeated at least 3 times in this study. Data were expressed as the mean ±standard deviation (SD). All statistical analyses were performed using GraphPad version 6.0 software (GraphPad Software Inc., La Jolla, CA, USA). Student’s t-test was used to analyze differences between two groups, and the significance of differences among multiple groups was analyzed by one-way analysis of variance (ANOVA) followed by Turkey’s post hoc test. P<0.05 was regarded as statistically significant.
Results
AGAP2-AS1 Expression Was Increased in Glioma Tissues and Cells
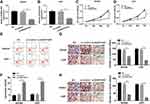
To explore the potential role of AGAP2-AS1 in glioma, the qRT-PCR was used to analyze the expression of AGAP2-AS1 in glioma tissues and cells. The results showed that AGAP2-AS1 expression was increased in LGG cases and HGG cases compared with that in NBTs. Moreover, HGG cases had higher AGAP2-AS1 expression than LGG cases (Figure 1A). Furthermore, the relative higher level of AGAP2-AS1 was observed in glioma cell lines (A172, LN229, SHG44 and U251) rather than in NHA cells, especially in SHG44 and U251 cells (Figure 1B). Thus, we chose SHG44 and U251 cells for further investigation. Furthermore, the relationship between the relative expression of AGAP2-AS1 and clinicopathologic characteristics of glioma patients was analyzed. As shown in Supplementary Table 1, the high expression level of AGAP2-AS1 was significantly associated with tumor size and WHO grade, but not with age, gender lymphatic metastasis, and differentiation grade. All these results indicated that AGAP2-AS1 might play an oncogenic role in glioma.
Knockdown of AGAP2-AS1 Suppressed Proliferation, Migration and Invasion, but Facilitated Apoptosis in Glioma Cells
To analyze the functional roles of AGAP2-AS1 in glioma, we interfered with endogenous AGAP2-AS1 expression in SHG44 and U251 cells via transfection with specific siRNA. The transfection efficiency was evaluated using qRT-PCR. As shown in Figure 2A and B, the expression of AGAP2-AS1 was dramatically decreased in SHG44 and U251 cells transfected with si-AGAP2-AS1 compared with those cells transfected with si-control or NC group (untransfected cells). CCK-8 assay suggested that knockdown of AGAP2-AS1 markedly impaired SHG44 and U251 cell growth (Figure 2C and D). Moreover, the change in the cell apoptosis after silencing AGAP2-AS1 was analyzed using flow cytometry analysis. As presented in Figure 2E and F, silenced AGAP2-AS1 induced the apoptosis of SHG44 and U251 cells. Transwell assay demonstrated that the depletion of AGAP2-AS led to a significant suppression of migration and invasion abilities in SHG44 and U251 cells (Figure 2G and H). All these findings proved that the AGAP2-AS1 was a tumor promoter in glioma progression in vitro.
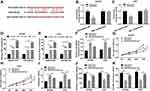
AGAP2-AS1 Served as a Molecular Sponge of miR-628-5p in Glioma Cells
It is generally accepted that lncRNAs could function as sponges or decoys of miRNAs to modulate target mRNAs expression. Thus, we intended to explore whether AGAP2-AS1 could modulate gene expression through sponging miRNAs. The starBase v3.0 was employed to search AGAP2-AS1 targets, miR-628-5p was found to potentially bind to AGAP2-AS1 (Figure 3A). Then, a dual-luciferase reporter system was used to determine whether miR-628-5p could directly bind to AGAP2-AS1. The results showed that overexpression of miR-628-5p remarkably decreased the luciferase activity of WT-AGAP2-AS1, whereas no obvious change on the luciferase activity of MUT-AGAP2-AS1 was found in SHG44 and U251 cells (Figure 3B and C). To determine whether AGAP2-AS1 and miR-628-5p were in the same RNA induced silencing complex (RISC) complex, RIP experiments were performed. The results showed that AGAP2-AS1 and miR-628-5p were greatly enriched in Ago2-containing beads compared with IgG control group (Figure 3D and E), suggesting the endogenous interaction between AGAP2-AS1 and miR-628-5p. Besides, the upregulation of miR-628-5p resulted in a significant promotion of miR-628-5p expression, while overexpression of AGAP2-AS1 attenuated the expression (Figure 3F). Moreover, restoration of miR-628-5p prominently blocked the proliferation, migration and invasion but induced apoptosis of SHG44 and U251 cells, while these effects were abolished by upregulating AGAP2-AS1 (Figure 3G–K). These results disclosed that AGAP2-AS1 might exert biological function via serving as a sponge of miR-628-5p in glioma cells.
PTN Was a Direct Target of miR-628-5p in Glioma Cells
To elucidate the underlying mechanism of miR-628-5p exerting its functional effects in glioma, the downstream targets of miR-628-5p were predicted by TargetScan. As shown in Figure 4A, putative binding sites between PTN and miR-628-5p, suggesting that PTN might be a target of miR-628-5p. To validate this prediction, we constructed the 3ʹUTR-WT-PTN or 3ʹUTR-MUT-PTN luciferase reporter vector and introduced them into SHG44 and U251 cells. Dual-luciferase reporter assay suggested that the luciferase activity was strongly suppressed in SHG44 and U251 cells co-transfected with 3ʹUTR-WT-PTN and miR-628-5p, while it was not changed in 3ʹUTR-MUT-PTN group (Figure 4B). In addition, the effect of miR-628-5p on the levels of PTN mRNA and protein in U251 and SHG44 cells was explored. The data showed that the mRNA and protein levels of PTN were decreased after transfection with miR-628-5p mimic (Figure 4C and D), suggesting PTN was negatively regulated by miR-628-5p. Moreover, PTN mRNA and protein levels were notably decreased in SHG44 and U251 cells transfected with si-PTN, while the effect was abated by co-transfection with miR-628-5p inhibitor (Figure 4E and F). Besides, downregulation of miR-628-5p partly reversed anti-proliferation, anti-migration, anti-invasion, and pro-apoptosis effects caused by PTN knockdown (Figure 4G–J). Taken together, these data indicated that miR-628-5p exerted its anti-tumor function via regulating PTN expression in glioma cells.
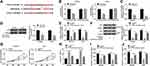
PTN Was Regulated by AGAP2-AS1 and miR-628-5p
To determine whether AGAP2-AS1 functioned as a ceRNA of miR-628-5p to regulate PTN expression, SHG44 and U251 cells were transfected with si-control, si-AGAP2-AS1, si-AGAP2-AS1 + inhibitor-control, or si-AGAP2-AS1 + miR-628-5p inhibitor. The qRT-PCR and Western blot assays showed that transfection with si-AGAP2-AS1 reduced the mRNA and protein expression of PTN in SHG44 and U251 cells, while the decreased expression of PTN was increased again by co-transfection with miR-628-5p inhibitor (Figure 5A–C). Collectively, our data revealed that AGAP2-AS1 functioned as a ceRNA of miR-628-5p to modulate PTN expression.
Knockdown of AGAP2-AS1 Inhibited Glioma Tumorigenesis in vivo
To confirm the effect of AGAP2-AS1 on glioma in vivo, U251 cells stably transfected with sh-NC or sh-AGAP2-AS1 were subcutaneously injected into the flanks of nude mice. Knockdown of AGAP2-AS1 greatly inhibited tumor volume and weight in xenograft model (Figure 6A and B). Additionally, qRT-PCR analysis presented that knockdown of AGAP2-AS1 led to a significant reduction of AGAP2-AS1 expression, while an increase of miR-628-5p level in excised tumor masses (Figure 6C and D). Furthermore, qRT-PCR and Western blot assays demonstrated that deficiency of AGAP2-AS1 evidently reduced the mRNA and protein expression of PTN (Figure 6E and F). Thus, it was concluded that AGAP2-AS1 inhibition reduced tumor growth by regulating miR-628-5p and PTN in vivo.
Discussion
Glioma is the most common and lethal brain tumor with an extremely poor prognosis. The aim of this research was to find a novel underlying mechanism in tumorigenesis and progression of glioma. Recently, increasing lncRNAs have been identified to be involved in the occurrence and development of different tumors, including glioma.22,23 Here, we focused on explaining the biological functions and the potential mechanism of AGAP2-AS1 in the occurrence and progression of glioma.
AGAP2-AS1 has been indicated to be dysregulated in many diseases and participated in multiple cell behaviors, such as proliferation, apoptosis, metastasis, and differentiation.12,24 Notably, AGAP2-AS1 was confirmed to promote cell proliferation in glioma cells via sponging miR-15a/b-5p to regulate HDGF.14 In addition, Tian et al proved that AGAP2-AS1 depletion limited cell growth and metastasis, but accelerated apoptosis in glioblastoma (GBM).25 Moreover, Luo et al demonstrated that AGAP2-AS1 level was enhanced in GBM and positively associated with poor prognosis, and AGAP2-AS1 depletion repressed growth and invasion but facilitated apoptosis in GBM cells through silencing TFPI2.26 Nevertheless, the regulatory mechanisms of AGAP2-AS1 in glioma are still not well understood. Firstly, we found that AGAP2-AS1 abundance was enhanced in HGG cases compared with NBTs or LGG cases, which was in line with previous studies. Subsequently, we characterized the functional roles of AGAP2-AS1 in glioma by using loss-of-function experiments. The resulted disclosed that AGAP2-AS1 inhibition suppressed the glioma cell growth and metastasis, and contributed to apoptosis. Similarly, in vivo experiments uncovered that AGAP2-AS1 depletion repressed tumor growth. Our findings combined with previous studies demonstrated that AGAP2-AS1 played an oncogenic role in the progression of glioma.
It is widely accepted that lncRNAs commonly modulate genes expression via sharing the same miRNAs with their downstream mRNAs.27 In this research, starBase 3.0 was employed to search for AGAP2-AS1 targets, and miR-628-5p was found to potentially bind to AGAP2-AS1. Then, dual-luciferase reporter assay suggested that AGAP2-AS1 directly targeted miR-628-5p. MiR-628-5p has been proven to be a tumor suppressor or promoter in different tumors. For example, miR-628-5p has been demonstrated to block the progression of epithelial ovarian cancer via inhibiting FGFR2.16 Additionally, Wang et al demonstrated that miR-628-5p level was elevated in osteosarcoma and its knockdown blocked the proliferation and metastasis of osteosarcoma cells through targeting IFI44L.28 Besides, Xie et al found that miR-628-5p repressed glioma cell growth via targeting DDX59.19 In the current research, we found that restoration of miR-628-5p obviously limited the cell growth, migration and invasion but facilitated apoptosis in glioma cells, while these effects were abolished by upregulating AGAP2-AS1. Our findings suggested that miR-628-5p acted as a tumor suppressor in glioma. To clarify how miR-628-5p affected the progression of glioma, potential target was predicted by TargetScan. PTN containing binding sites of miR-628-5p was selected as the target for further analysis.
The emerging evidence has suggested that PTN was abnormally expressed in various cancers.29,30 Previous studies demonstrated that PTN expression was enhanced in glioma and positively correlated with poor survival, and PTN accelerated migration and proliferation in glioma cells, while its knockdown inhibited tumor growth in vitro and in vivo.21,31,32 Consistent with these findings, we also proved that downregulation of PTN limited cell proliferation and metastasis but facilitated apoptosis in glioma cells, whereas co-transfection of miR-628-5p abolished these effects. Moreover, the results displayed that PTN expression was positively regulated by AGAP2-AS1 and negatively regulated by miR-628-5p. In addition, deficiency of AGAP2-AS1 resulted in significant promotion of miR-628-5p expression and reduction of PTN level in tissues from xenograft mice. Therefore, these findings disclosed that AGAP2-AS1 functioned as a ceRNA via sponging miR-628-5p to modulate PTN expression in glioma.
Conclusion
In conclusion, these findings showed that AGAP2-AS1 knockdown blocked the progression of glioma through functioning as ceRNA of miR-628-5p to regulate PTN expression. Hence, the AGAP2-AS1/miR-628-5p/PTN axis might be a promising therapeutic target for the treatment of glioma.
Acknowledgments
The authors would like to thank the participants in this study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi:10.1016/j.cancergen.2012.10.009
2. Laws ER, Parney IF, Huang W, et al. Survival following surgery and prognostic factors for recently diagnosed malignant glioma: data from the glioma outcomes project. J Neurosurg. 2003;99(3):467–473. doi:10.3171/jns.2003.99.3.0467
3. Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs. 2016;20(5):S2–S8. doi:10.1188/11.CJON.291-297
4. Thakkar JP, Dolecek TA, Horbinski C, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23(10):1985–1996. doi:10.1158/1055-9965.EPI-14-0275
5. Li J, Xuan Z, Liu C, Goodenberger ML, Jenkins RB. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013;14(9):18790–18808. doi:10.3390/ijms140918790
6. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10(1):38. doi:10.1186/1476-4598-10-38
7. Zhang X, Sun S, Pu JKS, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48(1):1–8. doi:10.1016/j.nbd.2012.06.004
8. Wu Q, Xiang S, Ma J, et al. Long non-coding RNA CASC 15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN 1A and ZEB 1. Mol Oncol. 2018;12(6):799–813. doi:10.1002/1878-0261.12187
9. Huang Y, Xiang B, Liu Y, et al. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi:10.1016/j.canlet.2018.08.024
10. Tsai M-C, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi:10.1158/0008-5472.CAN-10-2483
11. Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202. doi:10.1186/s13046-018-0875-3
12. Li W, Sun M, Zang C, et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7(5):e2225. doi:10.1038/cddis.2016.126
13. Qi F, Liu X, Wu H, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10(1):48. doi:10.1186/s13045-017-0420-4
14. Zheng Y, Lu S, Xu Y, et al. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530. doi:10.1016/j.ijbiomac.2019.01.121
15. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
16. Li M, Qian Z, Ma X, et al. MiR-628-5p decreases the tumorigenicity of epithelial ovarian cancer cells by targeting at FGFR2. Biochem Biophys Res Commun. 2018;495(2):2085–2091. doi:10.1016/j.bbrc.2017.12.049
17. Srivastava A, Goldberger H, Dimtchev A, et al. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumor Biol. 2014;35(5):4867–4873. doi:10.1007/s13277-014-1638-1
18. Favreau AJ, Sathyanarayana P. miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk Res. 2012;36(3):334–341. doi:10.1016/j.leukres.2011.09.027
19. Xie P, Wang Y, Liao Y, et al. MicroRNA‐628‐5p inhibits cell proliferation in glioma by targeting DDX59. J Cell Biochem. 2019;120(10):17293–17302. doi:10.1002/jcb.28991
20. Kurtz A, Schulte AM, Wellstein A. Pleiotrophin and midkine in normal development and tumor biology. Crit Rev Oncog. 1995;6(2):151–177.
21. Zhang L, Kundu S, Feenstra T, et al. Pleiotrophin promotes vascular abnormalization in gliomas and correlates with poor survival in patients with astrocytomas. Sci Signal. 2015;8(406):ra125–ra125. doi:10.1126/scisignal.aaa1690
22. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
23. Li J, Zhang M, An G, et al. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp Biol Med. 2016;241(6):644–649. doi:10.1177/1535370215622708
24. Liu Z, Wang Y, Wang L, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):194. doi:10.1186/s13046-019-1188-x
25. Tian Y, Zheng Y, Dong X. AGAP2‐AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem. 2019;120(6):9056–9062. doi:10.1002/jcb.28180
26. Luo W, Li X, Song Z, et al. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging (Albany NY). 2019;11(11):3811–3823. doi:10.18632/aging.102018
27. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi:10.1016/j.semcdb.2014.05.015
28. Wang JY, Lu SB, Wang JQ. miR-628-5p promotes the growth and migration of osteosarcoma by targeting IFI44L. Biochem Cell Biol. 2019. doi:10.1139/bcb-2019-0001
29. Feng Z, Gao S, Wu Y, et al. Lung cancer cell migration is regulated via repressing growth factor PTN/RPTP β/ζ signaling by menin. Oncogene. 2010;29(39):5416–5426. doi:10.1038/onc.2010.282
30. Riegel AT, Wellstein A. The potential role of the heparin-binding growth factor pleiotrophin in breast cancer. Breast Cancer Res Treat. 1994;31(2–3):309–314. doi:10.1007/bf00666163
31. Powers C, Aigner A, Stoica GE, et al. Pleiotrophin signaling through anaplastic lymphoma kinase is rate-limiting for glioblastoma growth. J Biol Chem. 2002;277(16):14153–14158. doi:10.1074/jbc.M112354200
32. Ulbricht U, Eckerich C, Fillbrandt R, et al. RNA interference targeting protein tyrosine phosphatase ζ/receptor‐type protein tyrosine phosphatase β suppresses glioblastoma growth in vitro and in vivo. J Neurochem. 2006;98(5):1497–1506. doi:10.1111/j.1471-4159.2006.04022.x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.