Back to Journals » Cancer Management and Research » Volume 11
Long Intergenic Non-Coding RNA 01121 Promotes Breast Cancer Cell Proliferation, Migration, and Invasion via the miR-150-5p/HMGA2 Axis
Authors Wang Z, Wang P, Cao L, Li F, Duan S, Yuan G, Xiao L, Guo L, Yin H, Xie D, Zhu J, Chen X, Zhang M
Received 9 September 2019
Accepted for publication 16 December 2019
Published 30 December 2019 Volume 2019:11 Pages 10859—10870
DOI https://doi.org/10.2147/CMAR.S230367
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Zhuolu Wang,1 Pinghu Wang,1 Lin Cao,1 Fucheng Li,1 Shenjia Duan,1 Guorong Yuan,1 Lixin Xiao,1 Lin Guo,1 Hong Yin,1 Duying Xie,1 Jing Zhu,1 Xingchu Chen,1 Mengqi Zhang2
1Department of Breast Surgery, Hunan Provincial Maternal and Child Health Care Hospital, Changsha 410008, People’s Republic of China; 2Department of Neurology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China
Correspondence: Xingchu Chen
Department of Breast Surgery, Hunan Provincial Maternal and Child Health Care Hospital, No. 53, Xiangchun Road, Kaifu District, Changsha 410008, People’s Republic of China
Tel +86-13974900511
Email [email protected]
Mengqi Zhang
Department of Neurology, Xiangya Hospital, Central South University, No. 87 Xiangya Road, Changsha 410008, People’s Republic of China
Tel +86-13548614858
Email [email protected]
Purpose: Long intergenic noncoding RNA 01121 (LINC01121) has been reported to be aberrantly expressed and acts as an oncogene in pancreatic cancer. However, the detailed molecular mechanism of LINC01121 in breast cancer remains largely unclear. In this study, we aimed to investigate the expression and biological function of LINC01121 in breast cancer.
Methods: LINC01121 and miR-150-5p expression were measured in breast cancer cell lines using quantitative reverse transcription PCR. MTS and flow cytometry assays were performed to determine cell proliferation, the cell cycle, and apoptosis. Cell migration and invasion were assessed by transwell assay. The protein expression of HMGA2 in breast cancer cell lines was measured by Western blotting. A luciferase reporter assay was used to assess the binding of LINC01121 and miR-150-5p.
Results: We found that LINC01121 was markedly up-regulated in breast cancer cell lines compared with normal breast epithelial cells. LINC01121 down-regulation markedly suppressed cell proliferation, cell cycle progression, migration, and invasion and promoted apoptosis in breast cancer cells. Further investigation showed that LINC01121 could serve as a molecular sponge for miR-150-5p and indirectly modulate the expression of its target, HMGA2. Moreover, miR-150-5p knockdown rescued the effects of LINC01121 down-regulation on HMGA2 protein expression, cell proliferation, cell cycle progression, apoptosis, migration, and invasion in breast cancer cells.
Conclusion: Knockdown LINC01121 inhibited breast cancer cell proliferation, migration, and invasion via the miR-150-5p/HMGA2 axis.
Keywords: breast cancer, LINC01121, growth, migration, invasion, miR-150-5p
Corrigendum for this paper has been published
Introduction
Breast cancer is the most commonly diagnosed invasive malignancy in females worldwide.1 In recent years, the incidence and mortality of breast cancer has been increasing.2 Epidemiological studies indicate that approximately 1.6 million cases of breast cancer are diagnosed and 1.2 million deaths result from breast cancer each year in China.3 During the past 10 years, many efforts have been made in breast cancer diagnosis and therapy, including combinations of surgery, hormone therapy, and adjuvant therapy.4 However, the overall prognosis of breast cancer is still unsatisfactory due to recurrence and distant metastasis.5 The proliferation and metastasis of breast cancer is a multi-step biological process that involves the activation or silencing of genes and epigenetic modifications; however, the details of its molecular mechanism are still largely unknown.6 Therefore, further elucidation of the molecular mechanisms of breast cancer metastasis is urgently needed to provide the theoretical basis for improved breast cancer therapy.
LncRNAs play key regulatory roles in various biological process, such as proliferation, invasion, and metastasis. They have been shown to be aberrantly expressed in many types of cancers, including breast cancer.7 HOTAIR has been shown to have increased levels of expression in breast cancer tissues and cells and may act as oncogene.8 Low levels of FENDRR expression are associated with decreased survival time of breast cancer patients and increased breast cancer cell proliferation and migration.9 However, the detailed mechanism of lncRNA-mediated tumorigenesis in breast cancer remains largely unclear. Long intergenic non-protein coding RNA 01121 (LINC01121) is a 1114 bp non-coding RNA located on human chromosome 2p21. LINC01121 has been reported to be aberrantly expressed and act as an oncogene in pancreatic cancer.10 Huang et al11 reported that pterostilbene could suppress proliferation and epithelial-to-mesenchymal transition and promote apoptosis in breast cancer cells. Further investigation showed that pterostilbene can decrease the expression of LINC01121, suggesting that LINC01121 may act as an oncogene in breast cancer. Therefore, we aimed to further study the function of LINC01121 in breast cancer.
LncRNAs act as competing endogenous RNAs (ceRNAs) to sponge microRNAs (miRNAs).12 miRNAs can affect various cellular processes by inhibiting the expression of their target genes.13,14 miR-150-5p, a hematopoiesis-related miRNA, has been frequently reported to be aberrantly expressed in various types of human cancers.15,16 Tang et al found that miR-150-5p has decreased levels of expression in breast cancer tissues and that miR-150 suppresses metastasis of breast cancer cells by inhibiting high-mobility group protein 2 (HMGA2) expression.17 However, whether LINC01121 acts as an miR-150-5p sponge to regulate the growth and metastasis of breast cancer cells remains unclear.
In the present study, we first evaluated the expression of LINC01121 in breast cancer cell lines. Subsequently, we assessed the biological function of LINC01121 in breast cancer cells. Finally, the interaction between LINC01121 and miR-150-5p and the underlying mechanisms of LINC01121 in breast cancer cells were investigated.
Materials and Methods
Cell Culture and Transfection
The human breast cancer cell lines, MCF-7, BT-549, MDA-MB-231, and MDA-MB-453 and the normal human breast cell line, MCF-10A, were purchased from the Shanghai Cell Bank of the Chinese Academy of Science (Shanghai, China). Cells were cultured in DMEM (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco) and incubated in a humidified incubator with 5% CO2 at 37 °C. An siRNAs against LINC01121 (si-LINC01121-): si-LINC01121-1(5ʹ-CAAAGACAAGGAUGAAAUAAG-3ʹ), si-LINC01121-2 (5ʹ-GACUAAACUAAUUAAGCUACA-3ʹ), si-LINC01121-3 (5ʹ-GUUCUCAUUUGAUGUUGAAUA-3ʹ), a negative control siRNA (si-NC, 5ʹ-UUCUCCGAACGUGUCACGUGC-3ʹ), miR-150-5p mimics (5ʹ-CTGGTACAGGCCTGGGGGACAG-3ʹ), a miR-150-5p inhibitor (5ʹ-CTGTCCCCCAGGCCTGTACCAG-3ʹ), a miRNA mimic negative control (NC mimic, 5ʹ-TTCTCCGAACGTGTCACGTAA-3ʹ), and a miRNA inhibitor negative control (NC inhibitor, 5ʹ-TTCTCCGAACGTGTCACGTAA-3ʹ) were purchased from GenePharma (Shanghai, China). Cell transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s protocol.
Nuclear and Cytoplasmic RNA Isolation
Cytoplasmic and nuclear RNAs were separated and purified using a PARIS kit (Thermo Fisher Scientific, Inc.). The expression of linc00467 in the nucleus and cytoplasm was measured by qRT-PCR.
Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). For LINC01121, first-strand cDNA was reverse transcribed from total RNA using the ImProm-IITM Reverse Transcription System (Promega, Madison, WI, USA). For miR-150-5p, cDNA was reverse transcribed using the ImProm-IITM Reverse Transcription System and stem-loop primer (Promega). The conditions of reverse transcription were 42°C for 60 min, followed by 85°C for 10 min. All PCRs were performed using an ABI 7500 RT-PCR system (Applied Biosystems) with SYBR® Premix Ex Taq™ Kit (TaKaRa, Kusatsu, Japan). PCR primers were purchased from GenePharma with the following sequences: LINC01121 forward, 5ʹ-TGGATGGATGGGTTGTGGTCTT-3ʹ and reverse, 5ʹ-TCCTTGTCTTTGTTACGCCTGT-3ʹ; GAPDH forward, 5ʹ-GCTCATTTGCAGGGGGGAG-3ʹ and reverse, 5ʹ- GTTGGTGGTGCAGGAGGCA-3ʹ; miR-150-5p forward, 5ʹ-ACACTCCAGCTGGG TCTCCCAACCCTTGTA-3ʹ and reverse, 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ; miR-1193 forward, 5ʹ- ACACTCCAGCTGGGATAGACCGGTGACGTGC-3ʹ and reverse, 5ʹ- CTCAACTGGTGTCGTGGA-3ʹ; miR-520h forward, 5ʹ- ACACTCCAGCTGGGAGTGCTTCCCTTTAGAG-3ʹ and reverse, 5ʹ- CTCAACTGGTGTCGTGGA-3ʹ; miR-520g-3p forward, 5ʹ- ACACTCCAGCTGGGATTCCCTTTAGAGTGT-3ʹ and reverse, 5ʹ- CTCAACTGGTGTCGTGGA-3ʹ; and U6 forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. GAPDH and U6 were used as endogenous controls for LINC01121 and miR-150-5p expression, respectively. Fold change in expression was calculated using the 2−ΔΔCT method.18 All experiments were repeated in independent triplicate.
MTS Assay
Cell proliferation was evaluated using a CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, Promega). In brief, after transfection, 96-well plates were seeded with 1 × 104 cells/100 μL/well in triplicate and incubated in a humidified incubator with 5% CO2 at 37 °C. The AQueous One Solution reagent (10 μL) was then added to the wells at 0, 24, 48, and 72 h. After cultured for 4 h at 37 °C, the absorbance was measured at 490 nm using a microplate reader (Bio-Rad, Hercules, CA, USA). All experiments were repeated in independent triplicate.
Cell Cycle and Apoptosis Assay
A Cell Cycle Detection Kit (Keygentec, Nanjing, China) was used to assess the cell cycle and an Annexin V-FITC Apoptosis Detection Kit (Keygentec) was used to detect apoptosis. The percentage of the cell population in different phases of the cell cycle and the percentage of the cells undergoing apoptosis were measured using flow cytometry (BD Biosciences, San Jose, CA, USA). All experiments were repeated in independent triplicate.
Transwell Migration and Invasion Assays
A BD 24-well transwell chamber (BD Biosciences) was used to assess cell migration and invasion. Briefly, 2 × 105 transfected MCF-7 or MD-MBA-231 cells, in 200 μL of serum-free medium, were seeded to the top chamber. For invasion assays, the upper chamber was coated in Matrigel (BD Biosciences), but for migration assays, Matrigel was not added. Medium containing 10% FBS was loaded into the lower transwell chamber. After incubation at 37 °C with 5% CO2 for 24 h, cells on the transwell membrane were stained with 0.5% crystal violet for 15 min. Migrant or invasive cells were counted under a microscope (Olympus, Tokyo, Japan). All experiments were repeated in independent triplicate.
Western Blotting
RIPA lysis buffer with protease inhibitor (Beyotime, Shanghai, China) was used to extract total protein. Equal amounts of denatured protein (30 μg) were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with Tris-buffered saline (TBS), containing 5% non-fat milk powder, at 25 °C for 2 h. The membranes were then incubated at 4 °C overnight with the following diluted primary antibodies: anti-HMGA2 (dilution 1:1000; ab97276, Abcam; Danvers, MA, USA) and anti-β-actin (dilution 1:1000; ab8227; Abcam). After washing with TBS containing Tween 20 (TBST), membranes were incubated with an HRP-labeled secondary antibody (dilution 1:5000) for 2 h at 25 °C. After washing three times with TBST, the protein bands were detected using an enhanced chemiluminescence detection kit (Beyotime) and a ChemiDoc™ XRS imaging system (Bio-Rad). All experiments were repeated in independent triplicate.
Luciferase Reporter Assays
Wild-type (WT) LINC01121 containing putative miR-150-5p binding sites and LINC01121 containing mutated miR-150-5p binding sites (MUT) were synthesized and then inserted into the luciferase reporter vector, psi-CHECK-2 (Promega). For luciferase assays, MCF-7 cells were co-transfected with luciferase reporter plasmids and an miR-150-5p mimic, an miR-150-5p inhibitor, or a negative control miRNA. Forty-eight hours after transfection, relative luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instructions. All experiments were repeated in independent triplicate.
Statistical Analysis
Statistical analyses were performed using SPSS 19.0 statistical software (IBM Inc., Chicago, IL, USA). Data are presented as means ± standard deviation (SD). A t-test was performed to detect significant differences between two groups. An ANOVA was performed to detect significant differences between three groups. A p-value less than 0.05 was regarded as statistically significant.
Results
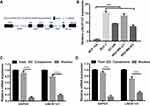
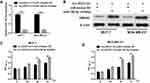
LINC01121 Was Markedly Up-Regulated in Breast Cancer Cell Lines
The structure of LINC01121 (1095bp) such as chromosomal locations and exons were diagrammed in Figure 1A. First, we investigated the relative expression levels of LINC01121 in breast cancer cell lines by qRT-PCR. The expression of LINC01121 in breast cancer cell lines (including MCF-7, BT-549, MDA-MB-231, and MDA-MB-453) was markedly up-regulated, especially in MCF-7 and MDA-MB-231 cell lines, compared with its expression in the normal breast epithelial cell line, MCF-10A (Figure 1B). Therefore, MCF-7 and MDA-MB-231 cell lines were selected for subsequent experiments. Additionally, we found that LINC01121 expression was significantly higher in cytoplasm than that in nuclear in MCF-7 and MDA-MB-231 cell lines (Figure 1C and D).
LINC01121 Down-Regulation Significantly Suppressed Breast Cancer Cell Proliferation and Cell Cycle Progression and Promoted Apoptosis
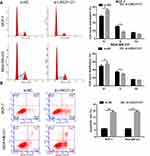
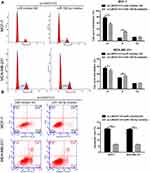
To investigate the biological function of LINC01121 in breast cancer cells, we transfected MCF-7 and MDA-MB-231 cells with si-LINC01121 to down-regulate LINC01121 expression. qRT-PCR results showed that the expression of LINC01121 was markedly decreased in both MCF-7 and MDA-MB-231 cells after transfected si- LINC01121-1/2/3, particularly si-LINC01121-1 which chose for further study (Figure 2A). MTS results showed that LINC01121 down-regulation significantly suppressed the proliferation of MCF-7 and MDA-MB-231 cells compared with cells transfected with si-NC (Figure 2B and C). Cell cycle analysis revealed that LINC01121 down-regulation increased the fraction of MCF-7 and MDA-MB-231 cells in G1 phase and decreased the fraction of MCF-7 and MDA-MB-231 cells in S phase compared with cells transfected with si-NC (Figure 3A). LINC01121 down-regulation significantly promoted the apoptosis of MCF-7 and MDA-MB-231 cells compared with cells transfected with si-NC (Figure 3B). These data suggested that LINC01121 down-regulation suppressed breast cancer cell proliferation and cell cycle progression and promoted apoptosis.
LINC01121 Down-Regulation Significantly Suppressed Breast Cancer Cell Migration and Invasion
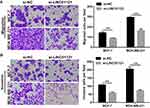
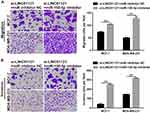
Next, transwell experiments showed that LINC01121 down-regulation significantly suppressed cell migration and invasion in both MCF-7 and MDA-MB-231 cells compared with cells transfected with si-NC (Figure 4).
LINC01121 Directly Bounds to miR-150-5p
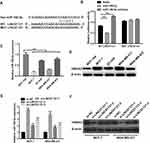
Next, we investigated whether LINC01121 acted as a ceRNA of miRNAs in breast cancer. Bioinformatics prediction analysis showed that LINC01121 has theoretical binding site for miR-1193, miR-520h, miR-520g-3p, and miR-150-5p. However, previous studies reported that only miR-150-5p is an anti-cancer gene.17 Thus, miR-150-5p was selected for further investigation. The theoretical binding site between LINC01121 and miR-150-5p was shown in Figure 5A. To verify that LINC01121 can directly bind to miR-150-5p, luciferase reporter assays were performed. Compared with the blank+ WT LINC01121 group, relative luciferase activity was inhibited in miR-150-5p+ WT LINC01121 group while it was promoted in miR-150-5p inhibitor+ WT LINC01121 group (Figure 5B). However, the relative luciferase activity was not affected between MUT LINC01121 +blank, MUT LINC01121 +miR-150-5p, and MUT LINC01121 +miR-150-5p inhibitor groups (Figure 5B). Next, miR-150-5p expression levels were significantly lower in the four breast cancer cell lines than that in the normal breast epithelial cell line, MCF-10A (Figure 5C). Previous studies have shown that miR-150-5p modulates breast cancer metastasis by targeting HMGA2.17 We found that HMGA2 expression was up-regulated in four breast cancer cell lines compared with its expression in MCF-10A cells (Figure 5D). Moreover, we found that miR-150-5p expression was promoted while HMGA2 expression was inhibited in MCF-7 and MDA-MB-231 cells after transfected si-LINC01121-1/2/3, particularly si-LINC01121-1 (Figure 5E–F). Additionally, other three predict miRNA (miR-1193, miR-520h, miR-520g-3p) expression after LINC01121 knockdown at 48 h were measured by qRT-PCR. The results showed that LINC01121 knockdown promoted miR-1193, miR-520h, and miR-520g-3p expression (Supplement Figure 1). But the change of miR-150-5p expression significantly higher than the change of other three miRNA (miR-1193, miR-520h, miR-520g-3p). These data suggested that LINC01121 may directly bound to miR-150-5p to regulate HMGA2 expression.
Knockdown miR-150-5p Significantly Attenuated the Effects of LINC01121 Down-Regulation on HMGA2 Protein Expression, Cell Proliferation, Cell Cycle Progression, and Apoptosis in Breast Cancer Cells
To further investigate the relationship between LINC01121 and miR-150-5p, we transfected MCF-7 and MDA-MB-231 cells that had previously been transfected with si-LINC01121, with an miR-150-5p inhibitor. miR-150-5p expression significantly decreased, while HMGA2 expression significantly increased, in MCF-7 and MDA-MB-231 cells co-transfected an miR-150-5p inhibitor and si-LINC01121 compared with cells co-transfected with an NC inhibitor and si-LINC01121 (Figure 6A and B). Moreover, miR-150-5p knockdown promoted cell proliferation (Figure 6C and D), increased the fraction of cells in S phase, decreased the fraction of cells in G1 phase (Figure 7A), and suppressed apoptosis (Figure 7B) in MCF-7 and MDA-MB-231 cells co-transfected with an miR-150-5p inhibitor and si-LINC01121 compared with cells co-transfected with an NC inhibitor and si-LINC01121. These data suggested that miR-150-5p knockdown attenuated the effects of si-LINC01121 on HMGA2 protein expression, cell proliferation, cell cycle progression, and apoptosis in MCF-7 and MDA-MB-231 cells.
miR-150-5p Knockdown Significantly Attenuated the Repressive Effects of LINC01121 Down-Regulation on the Migration and Invasion of Breast Cancer Cells
Transwell migration and invasion assays showed that miR-150-5p knockdown significantly promoted the migration and invasion of MCF-7 and MDA-MB-231 cells co-transfected an miR-150-5p inhibitor and si-LINC01121, compared with cells co-transfected with an NC inhibitor and si-LINC01121 (Figure 8).
Discussion
LncRNAs function as regulators of gene expression and have a great impact on the development and progression of various human cancers, including breast cancer.19 In the present study, we found that LINC01121 was markedly up-regulated in breast cancer cell lines compared with normal breast epithelial cells. LINC01121 down-regulation significantly suppressed cell proliferation, cell cycle progression, migration, and invasion and promoted apoptosis in MCF-7 and MDA-MB-231 cells. These results indicated that LINC01121 acts as an oncogene and is involved in the process of breast cancer proliferation, migration, and invasion. Qian et al10 reported that LINC01121 is overexpressed and acts as an oncogene in pancreatic cancer tissues, which is similar to our results.
One of the functions of lncRNAs is to act as miRNA sponges to regulate the expression of their miRNA targets. Herein, we used bioinformatics tools to predict that miR-150-5p was a target of LINC01121. The interaction between LINC01121 and miR-150-5p was confirmed using luciferase reporter assays, which showed that LINC01121 could directly bind to miR-150-5p. Moreover, LINC01121 knockdown promoted miR-150-5p expression in both MCF-7 and MDA-MB-231 cells. These results suggested that LINC01121 directly bound to miR-150-5p. Tang et al17 found that miR-150-5p suppresses the migration of breast cancer cells and its expression is decreased in triple-negative breast cancer tumor tissues, indicating that miR-150-5p may play an inhibitory role in triple-negative breast cancer. Hu et al20 found that BLACAT1 promotes breast cancer cell growth and metastasis by sponging miR-150-5p. Alipoor et al also found that MIAT knockdown suppresses MCF7 cell growth and migration by sponging miR-150-5p.21 Further rescue experiments demonstrated that miR-150-5p knockdown significantly reversed the effects of LINC01121 silencing on cell proliferation, cell cycle progression, apoptosis, migration, and invasion in MCF-7 and MDA-MB-231 cells. Taken together, these data indicated that LINC01121 down-regulation suppressed breast cancer cell proliferation and metastasis through the miR-150-5p axis, which is similar to the results of Hu and Alipoor.20,21
Previous studies have demonstrated that HMGA2 is highly overexpressed in breast cancer and promotes breast cancer cell proliferation and metastasis.22,23 miR-150-5p suppresses triple-negative breast cancer metastasis by inhibiting HMGA2 expression.17 In the present study, we found that HMGA2 was overexpressed in breast cancer cell lines, which is similar to the results of previous studies.17,22,23 Additionally, we found that LINC01121 knockdown inhibited HMGA2 expression in MCF-7 and MDA-MB-231 cells and that inhibiting miR-150-5p expression promoted HMGA2 expression and reversed the effect of LINC01121 silencing on HMGA2 expression. These data indicated that HMGA2, a target gene of miR-150-5p, is involved in the LINC01121-mediated regulation of breast cancer cell proliferation migration, and invasion.
Conclusion
LINC01121 knockdown suppressed breast cancer cell proliferation, migration, and invasion through the miR-150-5p/HMGA2 axis. These findings suggested that LINC01121 may serve as a novel molecular target for breast cancer therapy. We will confirm the effect of LINC01121 on breast cancer cell growth and metastasis in vitro in future experiments.
Abbreviations
LINC01121, Long intergenic noncoding RNA 01121; ceRNAs, competing endogenous RNAs; miRNAs, microRNAs; HMGA2, high-mobility group protein 2; qRT-PCR, quantitative reverse transcription PCR; SD, standard deviation.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81501025) and the Natural Science Foundation of Hunan Province (No. 2016JJ3174).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi:10.3322/caac.21412
3. Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–289. doi:10.1016/S1470-2045(13)70567-9
4. Ribeiro Pereira ACP, Koifman RJ, Bergmann A. Incidence and risk factors of lymphedema after breast cancer treatment: 10 years of follow-up. Breast. 2017;36:67–73. doi:10.1016/j.breast.2017.09.006
5. Lemler DJ, Lynch ML, Tesfay L, et al. DCYTB is a predictor of outcome in breast cancer that functions via iron-independent mechanisms. Breast Cancer Res. 2017;19(1):25. doi:10.1186/s13058-017-0814-9
6. Golse N, Adam R. Liver metastases from breast cancer: what role for surgery? Indications and results. Clin Breast Cancer. 2017;17(4):256–265. doi:10.1016/j.clbc.2016.12.012
7. Bai J, Zhao WY, Li WJ, Ying ZW, Jiang DQ. Long noncoding RNA LINC00473 indicates a poor prognosis of breast cancer and accelerates tumor carcinogenesis by competing endogenous sponging miR-497. Eur Rev Med Pharmacol Sci. 2019;23(8):3410–3420. doi:10.26355/eurrev_201904_17705
8. Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855. doi:10.1002/cam4.2018.7.issue-3
9. Li Y, Zhang W, Liu P, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–1412. doi:10.2147/OTT
10. Qian YG, Ye Z, Chen HY, et al. LINC01121 inhibits cell apoptosis while facilitating proliferation, migration, and invasion though negative regulation of the Camp/PKA signaling pathway via GLP1R. Cell Physiol Biochem. 2018;47(3):1007–1024. doi:10.1159/000490167
11. Huang Y, Du J, Mi Y, et al. Long non-coding RNAs contribute to the inhibition of proliferation and EMT by pterostilbene in human breast cancer. Front Oncol. 2018;8:629. doi:10.3389/fonc.2018.00629
12. Wang XB, Wang H, Long HQ, Li DY, Zheng X. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J Cell Physiol. 2019;234(5):7115–7127. doi:10.1002/jcp.27466
13. Cui J, Zhou B, Ross SA, Zempleni J. Nutrition, microRNAs, and human health. Adv Nutr. 2017;8(1):105–112. doi:10.3945/an.116.013839
14. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
15. Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. doi:10.1016/j.ccr.2012.08.028
16. Cao M, Hou D, Liang H, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50(5):1013–1024. doi:10.1016/j.ejca.2013.12.024
17. Tang W, Xu P, Wang H, et al. MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2. Onco Targets Ther. 2018;11:2319–2332. doi:10.2147/OTT.S161996
18. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi:10.1038/nprot.2008.73
19. Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed). 2015;7:94–108. doi:10.2741/s427
20. Hu X, Liu Y, Du Y, Cheng T, Xia W. Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 2019;9:14. doi:10.1186/s13578-019-0274-2
21. Alipoor FJ, Asadi MH, Torkzadeh-Mahani M. MIAT lncRNA is overexpressed in breast cancer and its inhibition triggers senescence and G1 arrest in MCF7 cell line. J Cell Biochem. 2018;119(8):6470–6481. doi:10.1002/jcb.v119.8
22. Sun M, Song CX, Huang H, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110(24):9920–9925. doi:10.1073/pnas.1305172110
23. Yang E, Cisowski J, Nguyen N, et al. Dysregulated protease activated receptor 1 (PAR1) promotes metastatic phenotype in breast cancer through HMGA2. Oncogene. 2016;35(12):1529–1540. doi:10.1038/onc.2015.217
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.