Back to Journals » OncoTargets and Therapy » Volume 14
LncRNA UCID Promotes Hepatocellular Carcinoma Metastasis via Stabilization of Snail
Authors Yuan S, Si W, Zhuang K, Li Y, Zhang Y, Liu J, Yang L, Zhang X
Received 20 August 2020
Accepted for publication 27 November 2020
Published 28 January 2021 Volume 2021:14 Pages 725—736
DOI https://doi.org/10.2147/OTT.S277951
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Shanshan Yuan,1,2,* Wangli Si,2,* Kun Zhuang,2 Yijun Li,2 Yanting Zhang,2 Jiaming Liu,2 Li Yang,3 Xin Zhang2
1Department of Gastroenterology, The Affiliated Xi’an Central Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 2Department of Gastroenterology, Xi’an Central Hospital, Xi’an, Shaanxi, People’s Republic of China; 3Department of Ultrasonography, The Affiliated Children’s Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li Yang
Department of Ultrasonography, The Affiliated Children’s Hospital of Xi’an Jiaotong University, No. 69, Xijuyuan Lane, Xi’an, Shaanxi, People’s Republic of China
Tel +86-29-87692073
Email [email protected]
Xin Zhang
Department of Gastroenterology, Xi’an Central Hospital, No. 161, Xiwu Road, Xi’an, Shaanxi, People’s Republic of China
Tel +86-29-62812284
Email [email protected]
Background: LncRNAs are functional regulators in tumor progression which act by regulating mRNAs in multiple types of cancer. However, the effect of lnc-UCID on hepatocellular carcinoma (HCC) metastasisremains unclear.
Methods: Lnc-UCID expression was quantified in HCC tissues and HCC cell lines by qRT-PCR. HCC cell lines with lnc-UCID knockdown were established by lentivirus transduction. The migration and invasion abilities of HCC cells were analyzed by Transwell and wound-healing assays. Protein expression of epithelial–mesenchymal transition (EMT)-related factors was examined by Western blot assay. Dual-luciferase assays and actinomycin D treatment were conducted to explore the relationship between lnc-UCID and Snail mRNA. The direct interaction between lnc-UCID and Snail mRNA was subjected to quantification analysis by biotinylated lnc-UCID pulldown assays. Pearson’s correlation coefficient was used to analyze correlations between lnc-UCID and Snail expression level in clinical samples. Rescue experiments were performed to uncover the role of Snail in the HCC metastasis process.
Results: Lnc-UCID was upregulated in human HCC tissues and HCC cell lines. Lnc-UCID promoted the cells’ mobility and invasiveness by enhancing the EMT process of HCC cells. The expression of Snail positively correlated with lnc-UCID abundance, and the interaction between lnc-UCID and Snail mRNA prevented miR-122, miR-203, miR-30b, miR-34a or miR-153 binding to the 3ʹ-UTR of Snail. Transfection of Snail greatly rescued the migration and invasion of HCC cells.
Conclusion: Lnc-UCID was upregulated in clinical HCC samples and directly interacted with Snail mRNA to enhance the stability of Snail mRNA, thus promoting the EMT process to accelerate HCC metastasis.
Keywords: hepatocellular carcinoma, HCC, long non-coding RNA, lnc-UCID, Snail, metastasis
Introduction
Hepatocellular carcinoma (HCC) ranks as the second most frequent cause of cancer-related deaths worldwide.1,2 The incidence and mortality of HCC have risen dramatically in the past 40 years, making it a major threat to human health.3,4 The median survival time of HCC is less than 1 year, mainly because most HCC patients are already in the advanced stage of HCC at the time of diagnosis. Moreover, tumor metastasis and a high rate of tumor recurrence also lead to the poor prognosis of HCC patients.1,3 Therefore, in order to reduce the incidence and mortality of HCC, there is an urgent need to explore effective therapeutic targets and strategies for HCC diagnosis and clinical treatment.
Long non-coding RNAs (LncRNAs) are a type of universally expressed non-coding RNA,5 participating in various physiological and pathological processes.6,7 LncRNAs regulate gene expression at the epigenetic, transcriptional and post-transcriptional levels, and also modify protein activities by directly interacting with proteins.8 Accumulated evidence shows that dysregulated expression of a variety of lncRNAs is closely correlated with cancer prognosis.9,10 Moreover, owing to the stability of lncRNAs, many cancer-related lncRNAs can function as non-invasive prognostic factors and therapeutic targets.11
Lnc-UCID is located at chromosome 1q22, and has been identified as an oncogenic factor in certain types of cancer. In colorectal cancer (CRC) cells, the upregulated lnc-UCID acts as an endogenous miRNA sponge, competing for miR-152-3p and thereby promoting CRC cell migration and invasion via the Wnt/β‑catenin signaling pathway.12 In HCC, the level of lnc-UCID is elevated and correlates with the shorter recurrence-free survival of HCC patients. Functional studies show that lnc-UCID enhances CDK6 expression by competitively binding to DHX9 and sequestering DHX9 from CDK6-3ʹ-UTR, thus promoting G1/S transition and hepatoma growth.13 However, the specific role of lnc-UCID and the related mechanism in the process of HCC metastasis remain unclear.
In this study, we found that lnc-UCID was highly expressed in HCC tissues and was associated with HCC metastasis. Mechanistic studies implied that lnc-UCID regulated Snail expression and enhanced the stability of Snail mRNA by preventing the inhibitory effect of miRNAs on Snail mRNA, thereby promoting the epithelial–mesenchymal transition (EMT) process and the metastasis of HCC cells. Our results deepen the understanding of the physiological role of lncRNAs in HCC metastasis, and have important theoretical significance for clinical application in the diagnosis and treatment of HCC.
Materials and Methods
Cell Culture and Clinical Tissue Samples
HCC tumor tissues and matched adjacent tissues were collected from patients who underwent surgical resection at Xi’an Central Hospital, and were stored at −80°C until analysis. The study was approved by the ethics committees of Xi’an Central Hospital, and written informed consent forms were signed by all subjects for research purposes. Human HCC cell lines, including HepG2, Huh7, Hep3B, HCCLM3 and SK-HEP-1, were obtained from either the American Type Culture Collection (Manassas, VA, USA) or the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, MA, USA) and penicillin–streptomycin antibiotics (Gibco, Thermo Fisher Scientific, MA, USA), and were incubated at 37°C with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, MA, USA) and reverse transcribed using PrimeScript™ (TaKaRa, Japan), and then subjected to quantification analysis by SYBR Premix Ex Taq™ (TaKaRa, Japan). The primer sequences were as follows: lnc-UCID, 5ʹ-CGGCCCACGGCAAAGAGA-3ʹ (forward), 5ʹ-TTGTACAGCCAGGTGTGGTG-3ʹ (reverse); Snail, 5ʹ- GCTGCAGGACTCTAATCCAGA-3ʹ (forward), 5ʹ-ATCTCCGGAGGTGGGATG-3ʹ (reverse). Relative expression of mRNA and miRNA was calculated with the 2−ΔΔCt method. Snail, lnc-UCID and other EMT-related factors were normalized to inner control GAPDH.
Cell Transfection and Lentiviral Transduction
For cell transfection, HCCLM3 and SK-HEP-1 cells were seeded at a density of 1×106 per well, and transfected with miRNAs, miRNA inhibitors or pcDNA3.0 (Thermo Fisher Scientific, MA, USA)-Snail plasmid and the corresponding negative controls using Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA). Short-hairpin oligonucleotides targeting lnc-UCID were synthesized and cloned into pLKO.1 construction (Gene Pharma, Shanghai, China). Lentiviral particles were packed based on pLKO.1-sh-lnc-UCID, psPAX2 (Gene Pharma, Shanghai, China) and pMD2.G (Gene Pharma, Shanghai, China) following common protocols. The shRNA sequences of lnc-UCID were: sh-lnc-UCID#1: 5ʹ-GAGCAAATTCAATGAGTAT-3ʹ (forward), 5ʹ-ATACTCATTGAATTTGCTC-3ʹ (reverse), sh-lnc-UCID#2:5ʹ-CTTCTGGCCTTGAGTGATT-3ʹ (forward), 5ʹ- AATCACTCAAGGCCAGAAG-3ʹ (reverse).
Transwell Migration and Invasion Assays
Transwell assays were performed according to the manufacturer’s guidelines. In brief, 2×104 transfected cells were suspended in 200 μL of serum-free medium and plated into the upper compartment of the chamber (Corning, NY, USA). The upper compartment was precoated with Matrigel (Sigma-Aldrich, MO, USA) for the invasion assays. After 24 h, the cells were stained with crystal violet and visually quantified.
Wound-Healing Assays
The HCC cells were seeded into 12-well plates at a density of 5×105 cells per well and grown until reaching 80% confluence. Then, the cell monolayer was linearly scratched with a sterile pipette tip. At 0 h and 48 h post-wounding, the cells were fixed with 4% paraformaldehyde for 15 min, followed by image recording to estimate the relative migration of cells. The quantification data were analyzed using ImageJ software version 1.41 (National Institutes of Health, MD, USA).
Luciferase Activity Assays
The promoter region of Snail was amplified and subcloned into the pGL3-Basic vector (Promega, WI, USA). HCCLM3 and SK-HEP-1 cells were seeded in 24-well plates at a density of 1×104 cells per well for 24 h before transfection. The cells were transfected with the indicated luciferase reporter plasmids, miRNA mimics and inner control Renila luciferase reporter vectors (pRL-TK) (Promega, WI, USA) using Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA) following the manufacturer’s protocol. Luciferase and Renilla signals were measured 48 h after transfection by a Dual-Luciferase Reporter Assay Kit (Promega, WI, USA). Luciferase activity was normalized to the corresponding Renilla luciferase values for the calculation of fold changes.
Biotinylated Lnc-UCID Pulldown Assays
The template DNA for in vitro transcription was produced by PCR to introduce a T7 promoter to the targeting region. Biotin-labeled lnc-UCID and its antisense RNA were transcribed in vitro based on the corresponding PCR product using T7 RNA polymerase (Roche, Mannheim, Germany) and Biotin RNA Labeling Mix (Roche, Mannheim, Germany). Then, the obtained product was purified and renatured with Annealing Buffer for RNA oligos (Beyotime, Shanghai, China) and heating at 90°C for 2 min, followed by incubation on ice for 2 min, and finally incubated at room temperature for 20 min. HCCLM3 and SK-HEP-1 cells were collected and lysed in the lysis buffer containing 30 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.5% NP-40, 60 U/mL RNase inhibitor (Promega, WI, USA) and protease inhibitor (Sigma-Aldrich, MO, USA). For the pulldown assay, 20 µL of Dynabeads M-280 Streptavidin beads (Thermo Fisher Scientific, MA, USA) were activated and blocked with 10 µg/mL RNase-free BSA and yeast tRNA (Sigma-Aldrich, MO, USA) for 30 min at 4°C. Cell lysates were incubated with the beads at room temperature for 2 h, followed by pulldown, RNA extraction and subsequent qRT-PCR quantification.
Western Blot Assay
Cells from a six-well plate were harvested and lysed using RIPA lysis buffer (Thermo Fisher Scientific, MA, USA). The primary antibodies against E-cadherin (Proteintech, IL, USA), N-cadherin (Proteintech, IL, USA), Vimentin (Proteintech, IL, USA), ZEB1 (Santa Cruz Biotechnology, Dallas, TX, USA), Snail (Santa Cruz Biotechonology, Dallas, TX, USA) and Twist (Proteintech, IL, USA) were used to interact with the targeting protein at 4°C overnight. After washing three times with 1×TBST, the membrane was incubated with HRP-labeled secondary antibodies (Jackson ImmunoResearch, PA, USA) for 1 h at room temperature. The blots were visualized with Pierce ECL Western Blotting chemiluminescence substrate (Thermo Fisher Scientific, MA, USA).
Statistical Analysis
SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. Specifically, the unpaired or paired Student’s t-test was used for comparison between two groups, the chi-squared test was utilized for the clinicopathological analysis, and Pearson’s correlation coefficient was used to analyze correlations between lnc-UCID and Snail expression level. The values are shown as the mean ± SD (standard deviation), and are representative of three independent experiments. **, ^^, P<0.01 indicates extremely significant differences and ns, P>0.05, indicates no significance.
Results
Lnc-UCID Level is Elevated in HCC and is Associated with Metastatic Characteristics
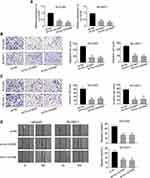
The expression levels of lnc-UCID in 80 pairs of HCC tumors and normal tissues were determined by qRT-PCR. The expression of lnc-UCID in HCC tumors was significantly higher than that of normal tissues (P<0.001) (Figure 1A). To confirm the results, we compared lnc-UCID abundance in HCC cell lines, including HepG2, Huh7, Hep3B, HCCLM3 and SK-HEP-1, and the normal liver epithelial cell line LO2. Similarly, lnc-UCID expression was greatly upregulated in HCC cell lines (P<0.01) (Figure 1B). To investigate the clinical significance of the high expression of lnc-UCID in HCC, the 80 enrolled patients were divided into the lnc-UCID low expression group (n=40) and lnc-UCID high expression group (n=40) according to the median value of lnc-UCID expression in HCC tissues (Figure 1A). The chi-squared test revealed that the expression level of lnc-UCID was closely related to HCC patients’ clinicopathological characteristics, including TNM stage, lymph-node metastasis, intrahepatic metastasis and distant metastasis (P<0.01), but not related to factors such as age, sex, tumor differentiation and tumor size (Table 1).
 |
Table 1 Correlation Between Lnc-UCID Expression and Clinicopathological Features of HCC Patients |
Knockdown of Lnc-UCID Dampens Metastasis of HCC Cells
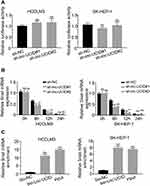
To understand the physiological role of lnc-UCID in HCC, we selected two HCC cell lines, HCCLM3 and SK-HEP-1, to establish stable lnc-UCID knockdown cell lines because of their highest lnc-UCID expression level. Compared with the sh-NC group, transduction of sh-lnc-UCID#1 or sh-lnc-UCID#2 effectively reduced lnc-UCID expression by more than 50% in the HCCLM3 and SK-HEP-1 cell lines (P<0.01) (Figure 2A). To study the effect of lnc-UCID on the migration and invasion of HCC cells, we performed Transwell experiments (without Matrigel) to evaluate the migration ability of HCCLM3 and SK-HEP-1 cells. The results showed that silencing of lnc-UCID significantly inhibited the migration of HCC cells (P<0.01) (Figure 2B). With regard to invasive ability, Transwell experiments (with Matrigel) showed that knockdown of lnc-UCID reduced the invasion capacity of HCCLM3 and SK-HEP-1 cells (P<0.01) (Figure 2C). In addition, the wound-healing assays further confirmed that lnc-UCID was critical for migration of HCC cells (Figure 2D). These results suggest that lnc-UCID promoted the metastasis of HCC cells in vitro.
Silencing of Lnc-UCID Inhibits the Epithelial–Mesenchymal Transition (EMT) Process
We observed that the HCC cells transduced with sh-lnc-UCID took on a round shape instead of the fusiform shape of a typical fibroblast, compared with sh-NC group (Figure 3A), suggesting that lnc-UCID may participate in the process of EMT. To confirm this hypothesis, we tested the effects of lnc-UCID on the expression of a series of EMT-related genes, including E-cadherin, N-cadherin, Vimentin, ZEB1, Snail and Twist, by qRT-PCR in HCCLM3 and SK-HEP-1 cells with different treatment. The results showed that knockdown of lnc-UCID increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin and Snail (P<0.01), but had no effect on mRNA levels of ZEB1 and Twist (Figure 3B). Western blot analysis also demonstrated that knockdown of lnc-UCID elevated the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin and Snail without affecting the expression of ZEB1 and Twist (Figure 3C).
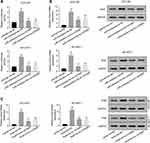
Lnc-UCID Interacts with Snail mRNA and Enhances Its Stability
The promoter region of Snail mRNA was cloned into the pGL3-basic vector, and the recombinant reporter plasmid was transfected into HCCLM3 and SK-HEP-1 cells, followed by fluorescence intensity measurements at 48 h post-transfection. Knockdown of lnc-UCID had no effect on Snail transcription (Figure 4A), indicating that lnc-UCID regulated Snail abundance at the post-transcriptional level. Then, actinomycin D was added to HCCLM3 and SK-HEP-1 cells, and the Snail expression level was determined by qRT-PCR at indicated time points. As shown in Figure 4B, knockdown of lnc-UCID significantly reduced the Snail mRNA level. To further investigate the interaction between lnc-UCID and Snail mRNA, a biotin-labeled lnc-UCID probe was used to perform RNA pulldown experiments in HCCLM3 and SK-HEP-1 cells. The qRT-PCR analysis revealed that the lnc-UCID probe enriched more Snail mRNA in HCC cells compared with the bio-NC group (Figure 4C). These experiments imply that lnc-UCID regulates the Snail level by interacting with Snail mRNA.
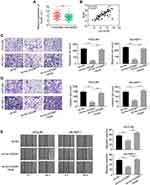
Association Between Lnc-UCID and Snail mRNA Blocks the Inhibitory Effects of miR-122, miR-203, miR-30b, miR-34a and miR-153 on Snail
Previous studies reported that miR-122, miR-203, miR-30b, miR-34a and miR-153 repressed Snail expression through binding to its 3ʹ-UTR.14–18 To probe whether the interaction between lnc-UCID and Snail affects the regulatory effects of these miRNAs on Snail, Snail 3′-UTR sequence was subcloned into the pmirGLO luciferase vector, and the reporter plasmids together with miR-NC or miRNAs inhibitors (cocktail containing inhibitors of miR-122, miR-203, miR-30b, miR-34a and miR-153) were co-transfected into HCCLM3 and SK-HEP-1 for dual-luciferase activity analysis. Inhibition of miR-122, miR-203, miR-30b, miR-34a and miR-153 significantly enhanced the luciferase activity, and knockdown of lnc-UCID strongly counteracted the enhancement effect of miRNAs on luciferase activity in HCC cells (Figure 5A and Supplementary Figure 1A). Similarly, transfection of inhibitors of miR-122, miR-203, miR-30b, miR-34a and miR-153 increased Snail expression, while simultaneously knockdown of lnc-UCID markedly abolished miRNA inhibition-mediated Snail upregulation in HCC cells (Figure 5B and Supplementary Figure 1B). A similar result was also obtained in Figure 5C. Compared with sh-NC-transduced HCC cells, silencing of lnc-UCID reduced Snail expression at both the mRNA and protein level.
Ectopic Expression of Snail Rescues Decreased Metastatic Ability of HCC Cells Caused by Lnc-UCID Knockdown
Quantification analysis of Snail mRNA in the 80 pairs of HCC tumors and normal tissues showed that the level of Snail was significantly increased in HCC tumors (P<0.001) (Figure 6A). Pearson’s correlation coefficient was exploited to analyze the correlation between lnc-UCID and Snail expression in the enrolled HCC patients. As shown in Figure 6B, lnc-UCID positively correlated with the expression of Snail mRNA abundance in HCC (P<0.001). To demonstrate the role of lnc-UCID and Snail interaction in HCC cell metastasis, Transwell experiments and wound-healing assays were performed to evaluate the migration and invasion abilities of HCCLM3 and SK-HEP-1 cells. Knockdown of lnc-UCID markedly suppressed the migration and invasion capacities of HCC cells (Figure 6C–E). However, simultaneous ectopic transfection of Snail obviously rescued the migration and invasion abilities of HCC cells transduced with sh-lnc-UCID (P<0.01).
Discussion
A number of dysregulated lncRNAs participate in a variety of biological processes in cancer, including occurrence, development and metastasis.19,20 In this study, with the qRT-PCR method, we found that long non-coding RNA UCID (lnc-UCID) was abnormally expressed in HCC tissues and cell lines, which is consistent with a previous study.13 Functional research revealed that upregulated lnc-UCID promoted the migration and invasion of HCC cells in vitro. Metastasis is the major reason for mortality and poor prognosis in HCC patients. Understanding the biological events and molecular mechanisms associated with HCC metastasis is of great significance for the effective diagnosis and clinical treatment of HCC. Combined with the accelerative role of lnc-UCID in HCC cell growth,13 our results implied that lnc-UCID plays a regulatory role in the malignant development of HCC. At present, there has been little research on the function of lnc-UCID in diseases. Previous studies suggested that lnc-UCID may have tissue or cancer type specificity, so the physiological effect of UCID in other types of cancers also needs to be explored.
In this study, we demonstrated that lnc-UCID increased the stability of Snail mRNA by interacting with Snail mRNA to prevent miRNA-mediated target mRNA degradation, thereby promoting the migration and invasion of HCC cells. EMT is a conserved process appearing at the invasive stage of metastatic cancer, which displays a loss of intracellular connections, reduced cell polarity and increased mesenchymal properties, thus enabling tumor cells migrate and invade to different tissues.21,22 EMT is not only correlated with tumor metastasis, but also closely associated with drug resistance in several types of cancer.23 We observed that lnc-UCID affected the EMT process of HCC cells and the expression of EMT-related factors, including E-cadherin, N-cadherin, Vimentin and Snail, suggesting that lnc-UCID is associated with the regulation of EMT in HCC cells. Therefore, whether lnc-UCID affects the sensitivity to chemical drug therapy of HCC is also worthy of further exploration.
Snail is a predominant inducer of EMT and its expression strongly correlates with tumor development and poor prognosis in a variety of tumors.24 As one transcriptional factor, Snail binds to the promoter region of E-cadherin and recruits the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex to initiate chromatin-modifying activities, thus repressing E-cadherin expression and promoting the EMT process in early development and in cancer progression.25,26 In the present study, we noticed that the level of Snail was upregulated in HCC tissues and positively correlated with the abundance of lnc-UCID. It has been reported that lncRNAs can regulate the EMT process of HCC. For examples, ectopic expression of lncRNA GAS5 elevates the E-cadherin level in HCC cells, which promotes the EMT process.27 LncRNA ATB competitively binds to the miR-200 family, thereby increasing the expression of ZEB1 and ZEB2 in HCC cells.28,29 In addition, HOTAIR acts as an miR-23b-3p sponge to positively regulate ZEB1 expression to enhance the process of EMT.30 Numerous studies in the literature have proved that lncRNAs can modulate gene expression at post-transcriptional levels through alternative splicing, acting as miRNA sponges, precursors of miRNAs, mediating RNA decay or maintaining mRNA stability. In HCCLM3 and SK-HEP-1 cells, lnc-UCID had no effect on Snail transcription, but altered Snail abundance at post-transcriptional level by maintaining the stability of Snail mRNA. The direct interaction between lnc-UCID and Snail mRNA prevents miR-122,14 miR-203,15 miR-30b,17 miR-34a16 and miR-15318 mediated inhibition of Snail mRNA expression, which is similar to a previous study.31 Therefore, we propose a regulatory model in which the regulatory effect of lnc-UCID on the EMT signaling pathway, especially on E-cadherin, is achieved by maintaining the stability of Snail mRNA, which further promotes the metastasis of HCC cells.
In conclusion, our findings unravel a novel regulatory mechanism of lnc-UCID in the process of HCC metastasis. The upregulated lnc-UCID binds to Snail to form a duplex, which prevents the binding of miRNAs on 3ʹ-UTR of Snail mRNA to maintain the stability of Snail, thus promoting the EMT process and metastasis of HCC cells.
Abbreviations
lncRNAs, long non-coding RNAs; HCC, hepatocellular carcinoma; miRNA, microRNA; EMT, epithelial–mesenchymal transition.
Data Sharing Statement
The data sets used during the present study are available from the corresponding author Li Yang on reasonable request.
Ethics Approval and Consent to Participate
The present study complied with the Declaration of Helsinki and was approved by the Ethical and Scientific Committee of Xi’an Central Hospital. The liver tumor and adjacent tissue samples were collected from patients undergoing surgical resection from February 2016 to August 2019. After obtaining the patient’s informed consent, the surgically resected samples were stored at −80°C for further analysis.
Acknowledgment
This study was supported by Xi’an Municipal Health Bureau (2017113SF/YX007-10).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi:10.3322/caac.20138
3. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi:10.1056/NEJMra1001683
4. Ryerson AB, Eheman CR, Altekruse SF, et al. Annual Report to the Nation on the Status of Cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi:10.1002/cncr.29936
5. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
6. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi:10.1016/j.canlet.2013.06.013
7. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi:10.1016/j.cell.2009.02.006
8. Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–116. doi:10.1093/bib/bbv031
9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
10. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
11. Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–1933. doi:10.1111/cas.13342
12. Sun LB, Zhao SF, Zhu JJ, Han Y, Shan TD. Long noncoding RNA UCID sponges miR1523p to promote colorectal cancer cell migration and invasion via the Wnt/betacatenin signaling pathway. Oncol Rep. 2020;44(3):1194–1205. doi:10.3892/or.2020.7670
13. Wang YL, Liu JY, Yang JE, et al. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology. 2019;70(1):259–275. doi:10.1002/hep.30613
14. Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp Cell Res. 2017;360(2):210–217. doi:10.1016/j.yexcr.2017.09.010
15. Zhang Z, Zhang B, Li W, et al. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes Cancer. 2011;2(8):782–791. doi:10.1177/1947601911429743
16. Dong P, Xiong Y, Watari H, et al. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer Res. 2016;35(1):132. doi:10.1186/s13046-016-0415-y
17. Xiong Y, Wang Y, Wang L, et al. MicroRNA-30b targets Snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. J Cancer. 2018;9(12):2147–2159. doi:10.7150/jca.25006
18. Xia W, Ma X, Li X, et al. miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting Snail. Oncol Rep. 2015;34(2):655–662. doi:10.3892/or.2015.4008
19. Wei L, Wang X, Lv L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18(1):147. doi:10.1186/s12943-019-1086-z
20. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77. doi:10.1186/s12943-020-01188-4
21. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi:10.1016/j.cell.2011.09.024
22. Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi:10.1172/JCI38019
23. Shintani Y, Okimura A, Sato K, et al. Epithelial to mesenchymal transition is a determinant of sensitivity to chemoradiotherapy in non-small cell lung cancer. Ann Thorac Surg. 2011;92(5):
24. Chen WJ, Wang H, Tang Y, Liu CL, Li HL, Li WT. Multidrug resistance in breast cancer cells during epithelial-mesenchymal transition is modulated by breast cancer resistant protein. Chin J Cancer. 2010;29(2):151–157. doi:10.5732/cjc.009.10447
25. Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–319. doi:10.1128/MCB.24.1.306-319.2004
26. Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi:10.1038/35000034
27. Chang L, Li C, Lan T, et al. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13(2):1541–1550. doi:10.3892/mmr.2015.4716
28. Li Y, Ye Y, Chen H. Astragaloside IV inhibits cell migration and viability of hepatocellular carcinoma cells via suppressing long noncoding RNA ATB. Biomed Pharmacother. 2018;99:134–141. doi:10.1016/j.biopha.2017.12.108
29. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010
30. Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–122. doi:10.1016/j.gene.2018.05.061
31. Wang GQ, Wang Y, Xiong Y, et al. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016;6:21865. doi:10.1038/srep21865
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.