Back to Journals » Cancer Management and Research » Volume 14
lncRNA SLC9A3-AS1 Promotes Oncogenesis of NSCLC via Sponging microRNA-760 and May Serve as a Prognosis Predictor of NSCLC Patients
Authors Huang X, Huang M, Chen M, Chen X
Received 3 December 2021
Accepted for publication 27 January 2022
Published 9 March 2022 Volume 2022:14 Pages 1087—1098
DOI https://doi.org/10.2147/CMAR.S352308
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Xiuming Huang,* Mingfang Huang,* Minbiao Chen, Xianshan Chen
Department of Thoracic Surgery, Hainan General Hospital, Haikou, Hainan, 570311, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xianshan Chen, Department of Thoracic Surgery, Hainan General Hospital, No. 19 Xiuhua Road, Haikou, Hainan, 570311, People’s Republic of China, Email [email protected]
Background: Non-small cell lung cancer (NSCLC) is a prevalent type of lung cancer worldwide. Long noncoding RNA (lncRNA) SLC9A3-AS1 is reported to play a carcinogenic role in nasopharyngeal carcinoma, but its full-scale role in NSCLC remains elusive.
Methods: SLC9A3-AS1 expression was detected in serum and tissue of NSLCC patients and NSCLC cell lines. The effects of SLC9A3-AS1 on NSCLC proliferation, migration and invasion were evaluated using CCK-8 and transwell assays. In addition, the potential downstream molecules of SLC9A3-AS1 were searched and explored by bioinformatics analysis, RT-qPCR, dual-luciferase reporter, and rescue experiments.
Results: SLC9A3-AS1 was upregulated in NSCLC tissues and cell lines. SLC9A3-AS1 possessed a favorable ability in diagnosing NSCLC. A high level of SLC9A3-AS1 was associated with poor prognosis in NSCLC patients. Functionally, SLC9A3-AS1 knockdown inhibited cell proliferation, migration, and invasion of NSCLC cells. Mechanistically, SLC9A3-AS1 acted as competing endogenous RNA for miR-760 to regulate NSCLC progression. In addition, rescue assay showed that downregulation of miR-760 could reverse the modulatory activity of SLC9A3-AS1 knockdown on NSCLC cells.
Conclusion: SLC9A3-AS1 was upregulated in NSCLC, and SLC9A3-AS1 knockdown hindered NSCLC progression through targeting miR-760, suggesting that it may prove to be a novel biomarker and therapeutic target for NSCLC.
Keywords: SLC9A3-AS1, NSCLC, miR-760, diagnosis, prognosis
Introduction
Lung cancer (LC) is the leading cause of cancer-related mortality worldwide.1 Non-small cell lung cancer (NSCLC) is the most common histological type of LC, accounting for about 80% of all LC cases and causing 28.49 death per 100,000 persons in China in 2014.2,3 Despite the remarkable therapeutic advances in surgical resection, chemotherapy, radiation therapy and immunotherapy, more than 75% LC patients were diagnosed at more advanced stages in China with a five-year survival rate of 15.6%.4–6 Despite the complex mechanisms of NSCLC oncogenesis and progression, molecular targeted therapy has developed into a promising breakthrough in the treatment of lung cancer.7 And, it is imperative to investigate the molecular mechanisms of NSCLC tumorigenesis and develop novel therapeutical target for NSCLC patients.
Long noncoding RNAs (lncRNAs) is a kind of non-translating RNAs with over 200 nucleotides in length. It is reported that aberrant expression of lncRNAs can be used as a prognosis predictor and therapeutic biomarkers in various human cancers.8–10 LncRNAs can exhibit the oncogenic or tumor-suppressive function via acting as competing endogenous RNAs (ceRNAs) by competitively binding to miRNA response elements.11 LncRNAs can be stably expressed in serum and other biological fluids with an appropriate and measurable concentration, which may provide valuable and cost-effective noninvasive biomarkers for patients with NSCLC.12,13 SLC9A3 antisense RNA 1 (SLC9A3-AS1) was first reported to be upregulated in extracellular vesicles from the peripheral blood of LC patients, which could be used to discriminate LC patients.14 Recently, Li et al reported that lncRNA SLC9A3-AS1 promoted the oncogenesis of nasopharyngeal carcinoma by modulating the miR-486-5p/E2F transcription factor 6 axis.15 Given the upregulation and discrimination power of SLC9A3-AS1 in LC patients, we further speculated that SLC9A3-AS1 might play a central role during the oncogenesis of NSCLC.
In this study, we intended to evaluate the diagnostic and prognostic abilities of SLC9A3-AS1 by measuring its expression in NSCLC and clarify the biological roles and downstream mechanisms of SLC9A3-AS1 in NSCLC.
Materials and Methods
Patients
This study was approved by the Ethics Committee of Hainan General Hospital (Haikou, China; Approval No. 20200012), and all the experimental procedures were performed in accordance with the Declaration of Helsinki. All enrolled individuals provided signed informed consents before initiation of the study. Included in this study were 130 consecutive patients with single pulmonary nodules (< 5 cm in diameter) determined by CT/LDCT scan who received treatment in our hospital between January 2016 and December 2020. All the included patients aged over 18 years and had no prior history of malignancies. They all underwent surgical resection and the diagnosis was confirmed by postoperative pathology. Among the 130 patients, 86 were confirmed as having NSCLC and the other 44 as having lung benign lesions (LBLs). In addition, a group of 40 age and sex-matched healthy controls (HC) were enrolled. The patients’ baseline characteristics are shown in Table 1. Blood samples were collected one day before surgery for patients with pulmonary nodules from the healthy controls during physical examinations. The peripheral blood samples were collected from all participants in serum gel separator tubes. Each blood sample was immediately centrifuged at 3000 g for 10 min at 4°C to separate sera, which were then stored at −80°C for use.
 |
Table 1 Patients Clinical Characteristics |
Cell Culture
Human normal lung epithelial cells (16HBE) and NSCLC cell lines (A549, H520, and H1299) were obtained from American Type Culture Collection (ATCC). All the cells were cultured in RPMI-1640 culture medium (Thermo Fisher Scientific, USA) containing 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA) in a 37 ◦C incubator with 5% CO2.
RNA Extraction and Reverse Transcription by Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and miRNA was isolated using a mirVana™ miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was then synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA) for SLC9A3-AS1 and a miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech Co., Ltd.) for miR-760 in accordance with the manufacturer’s protocol. RT-qPCR was then conducted using the SYBR® Premix Ex Taq (Takara Bio, Inc.) for SLC9A3-AS1 and a miRcute miRNA qPCR Detection Kit SYBR-Green (Tiangen Biotech Co., Ltd.) for miR-760. The relative expression levels of SLC9A3-AS1 and miR-760 were normalized to GAPDH and U6 using the 2−ΔΔCt method.16 All sequences of the oligonucleotides (GenePharma Co., Ltd, Shanghai, China) used in this study are shown in Table S1.
Cell Transfection
NSCLC cells were transfected with siRNAs against SLC9A3-AS1 (si-SLC9A3-AS1#1 or #2) and its negative control (siNC), miR-760 inhibitor (GenePharma Co., Ltd, Shanghai, China). In brief, NSCLC cells were seeded in 6-well plates at a density of 1×105 cells per well for 24h. Cell transfection was then conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) based on the manufacturer’s instructions. Transfected cells were cultured for an additional 48 h at 37°C before being used in downstream experiments.
Cell Counting Kit-8 (CCK-8) Proliferation Assays
Three replicates of transfected NSCLC cells were seeded into 96-well plates at a density of 1×104 cells per well. Then, 10 μL CCK-8 solution (Dojindo, Kumamoto, Japan) was added to the medium every 24 h until 72 h before the cells were further incubated at 37°C for 2 h. Optical density (OD) values were measured at 450 nm using an automatic microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell Assays
The migration and invasion abilities of NSCLC cells were detected by polyethylene membranes (24-well inserts; 8.0 μm; Corning, Inc.). The chambers precoated with 50 µL Matrigel (BD Biosciences) at 37°C for 1 h were used for invasion assay, and those uncoated chambers were used for migration assay. The upper chamber was seeded with cell suspension containing 1×105 cells in 100 μL FBS-free RPMI 1640, and the lower chamber was covered with 500 μL RPMI 1640 supplemented with 10% FBS. Cells were cultured at 5% CO2 and 37°C for 48 h. Cells that remained in the upper membrane were gently removed using a cotton swab, and cells that had migrated or invaded the bottom of the membrane were fixed with polyoxymethylene at room temperature for 20 min and then stained with 0.5% crystal violet at room temperature for 20 min. Cells were counted in five randomly selected fields under the light microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Subcellular Fractionation Assay
The nuclear and cytoplasmic fractions of NSCLC cells were separated using a PARIS kit (Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Next, total RNA was isolated from each fraction to quantify SLC9A3-AS1 expression in the nuclear and cytoplasmic fractions using RT-qPCR.
RNA Immunoprecipitation (RIP) Assay
The RIP assay was conducted using the EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer’s instructions. After centrifugation at 1000 x g for 10 min at 4°C, 10% cell lysate was used as the positive control (input). Human anti-Ago2 or IgG antibody (Millipore, USA) was utilized to capture the RNAs for RT-qPCR analysis. SLC9A3-AS1 or miR-760 expression was measured by RT-qPCR.
Luciferase Reporter Assay
The SLC9A3-AS1 fragments harboring the predicted wild-type (WT) or mutant (MUT) binding site (Shanghai GenePharma Co., Ltd.) were cloned into the pmirGLO luciferase plasmid (Promega Corporation) to create the reporter plasmids WT-SLC9A3-AS1 or MUT-SLC9A3-AS1. NSCLC cells were placed in a 24-well plate and co-transfected with WT- or MUT-SLC9A3-AS1, together with miR-760 mimic or NC-mimic using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48-h co-transfection, reporter activities were evaluated with a dual-luciferase reporter assay system (Promega Corporation, USA).
Statistical Analyses
Continuous variables are expressed as the mean ± standard deviation (SD), and categorical variables are expressed as frequencies with percentages. Differences between two groups were analyzed using the unpaired Student’s t-test. Comparisons of multiple groups were analyzed by analysis of variance (ANOVA) followed by Dunnett’s test. Categorical data were compared using the chi-squared test. Overall survival (OS) time was calculated from the date of initial surgery to the date of death or last follow-up. The recurrence-free survival (RFS) time was calculated from the date of initial surgery to the date of recurrence, death, or last follow-up. Kaplan-Meier curves were plotted for OS and RFS, and compared using Log rank test. Factors associated with OS or RFS were assessed by both univariate and multivariate Cox regression analyses using a forward stepwise approach. Prognostic factors with statistical significance (P<0.05) after univariate analysis were included in the Cox proportional hazards regression model for multivariate analysis. Receiver operating characteristic curves (ROC) were utilized to calculate diagnostic accuracy. Correlation analysis was performed using Pearson’s method. Statistical evaluations were performed using SPSS 20.0 (IBM SPSS Inc., Chicago, IL, USA). Differences were considered statistically significant when P< 0.05.
Results
SLC9A3-AS1 is Upregulated in NSCLC and Correlates with Advanced Characteristics
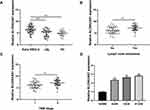
First, we investigated the expression profiles of SLC9A3-AS1 in NSCLC by RT-qPCR and found that the SLC9A3-AS1 expression level in NSCLC was significantly higher than that in LBL or HC (P< 0.001) (Figure 1A). In addition, SLC9A3-AS1 was expressed at an even higher level in NSCLC with lymph node metastasis (LNM) (P< 0.01) (Figure 1B) or at an advanced TNM stage (P< 0.01) (Figure 1C). Subsequently, the relative expression level of SLC9A3-AS1 was evaluated in human normal lung epithelial cells (16HBE) and NSCLC A549, H520 and H1299 cells. RT-qPCR data showed that SLC9A3-AS1 expression NSCLC cells was significantly higher than that in 16HBE cells (P< 0.01) (Figure 1D).
The Diagnostic and Prognostic Performance of Serum SLC9A3-AS1 in NSCLC Patients
First, we analyzed the performance of serum SLC9A3-AS1 in distinguishing NSCLC patients from LBL or HC using ROC analysis. As shown in Figure 2A, serum SLC9A3-AS1 proved to be a promising biomarker for distinguishing NSCLC patients from LBL, with an area under the curve (AUC) of 0.74 (95% CI: 0.65–0.82, P < 0.001). In addition, serum SLC9A3-AS1 exhibited a remarkable ability in differentiating NSCLC patients from HC, with an AUC of 0.79 (95% CI, 0.71–0.87, P < 0.001) (Figure 2B).
Next, we analyzed the relationship between SLC9A3-AS1 expression level in NSCLC tissues and the clinicopathological features of these NSCLC patients. First, we categorized the 86 NSCLC patients into a SLC9A3-AS1 low expression group (n=43) and a SLC9A3-AS1 high expression group (n=43) based on the median value of SLC9A3-AS1 expression in NSCLC tissues. It was found that the expression level of SLC9A3-AS1 was significantly correlated with LNM and TNM stage (Table 2). Next, the Kaplan-Meier curves indicated that higher expression level of SLC9A3-AS1 was associated with shorter OS (P= 0.033) (Figure 2C) and RFS (P= 0.031) (Figure 2D). Univariate and multivariate COX regression analyses in Tables 3 and 4 demonstrated that TNM stage, LNM and high SLC9A3-AS1 expression were independent indicators of poor OS and RFS in NSCLC patients. Collectively, our data indicate that SLC9A3-AS1 is an independent diagnostic and prognostic factor in NSCLC patients.
 |
Table 2 The Association Between SLC9A3-AS1 Expression and Clinicopathological Characteristics of NSCLC |
 |
Table 3 Univariate and Multivariate Cox Proportional Analyses of Prognostic Factors Associated with Overall Survival in NSCLC Patients |
 |
Table 4 Univariate and Multivariate Cox Proportional Analyses of Prognostic Factors Associated with Recurrence-Free Survival in NSCLC Patients |
SLC9A3-AS1 Knockdown Suppresses NSCLC Cell Proliferation, Migration and Invasion
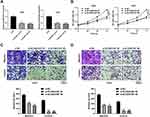
Next, we investigated the impact of SLC9A3-AS1 expression on NSCLC tumor progression in terms of cell proliferation, migration and invasion using CCK-8 and Transwell assays. First, we knocked down the expression of SLC9A3-AS1 in both A549 and H520 cells using two SLC9A3-AS1 siRNAs (P< 0.001) (Figure 3A). The results showed that silencing SLC9A3-AS1 expression significantly reduced the cell proliferation ability in both A549 and H520 (P< 0.01) (Figure 3B). Transwell assays demonstrated that downregulation of SLC9A3-AS1 reduced the migratory and invasive capabilities of the above two cell lines (P< 0.01) (Figure 3C and D).
SLC9A3-AS1 Serves as a Sponge for miR-760
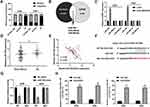
lncRNAs are known to have impact of miRNA expression on ceRNA identification.17 Our subcellular distribution assay showed that SLC9A3-AS1 was cytoplasmically located in NSCLC (Figure 4A), suggesting that it may serve as a miRNA sponge. After overlapping the putative target miRNAs of SLC9A3-AS1 using starBase and miRDB, three miRNAs (miR-760, miR-486-5p and miR-506-5p) were identified (Figure 4B). RT-qPCR further revealed that SLC9A3-AS1 knockdown increased the miR-760 expression but exerted no effect on the level of miR-486-5p and miR-506-5p (Figure 4C). Compared with HC, miR-760 was significantly downregulated in NSCLC (P< 0.01) (Figure 4D). Moreover, Pearson correlation analysis showed an inverse correlation between the expression of miR-760 and SLC9A3-AS1 in NSCLC (r= 0.507, P< 0.001) (Figure 4E). As demonstrated in Figure 4F, miR-760 contained sequences complementary to SLC9A3-AS1. To further determine the interaction of miR-760 and SLC9A3-AS1, we conducted the dual-luciferase reporter and RIP assays, showing that that the luciferase activity of NSCLC cells transfected with WT SLC9A3-AS1 and miR-760 mimic but not MUT SLC9A3-AS1 decreased significantly (P< 0.01) (Figure 4G). The data of RIP revealed a significant enrichment of SLC9A3-AS1 and miR-760 in NSCLC cells (P< 0.001) (Figure 4H). All these results demonstrated that SLC9A3-AS1 sponged miR-760 in NSCLC.
SLC9A3-AS1 Knockdown Inhibits NSCLC Cell Proliferation, Migration and Invasion via Sponging miR-760
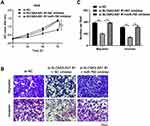
To determine whether miR-760 was involved in si-SLC9A3-AS1-mediated inhibition of malignancy in NSCLC, we transfected miR-760 inhibitor in SLC9A3-AS1 knockdown-A549 cells. As demonstrated in Figure 5A, miR-760 inhibition reversed the suppressive effect of si-SLC9A3-AS1 on NSCLC cell proliferation. Similarly, SLC9A3-AS1 knockdown alone distinctly impaired the migration and invasion abilities of A549 cells, while these impacts were obviously abolished after downregulation of miR-760 (Figure 5B and C). All these data showed that SLC9A3-AS1 knockdown inhibited NSCLC cell proliferation, migration and invasion by interacting with miR-760.
Discussion
Extensive lncRNAs have been recognized to exert critical biological functions in oncogenesis and progression of NSCLC.18 For instance, upregulation of SNHG14 facilitates NSCLC progression through targeting miR-206/G6PD pathway.19 Yu et al reported that overexpression of TMPO-AS1 could promote the malignant phenotypes of NSCLC via modulating miR-204-3p/ERBB2 axis.20 However, most lncRNAs, including SLC9A3-AS1, have not been addressed in the context of NSCLC. The present study demonstrated that SLC9A3-AS1 could serve as a biomarker for the diagnosis and prognostic prediction in patients with NSCLC. In addition, SLC9A3-AS1 was overexpressed in NSCLC, and downregulation of SLC9A3-AS1 hindered the progression of NSCLC via regulating miR-760.
Recently, SLC9A3-AS1 has been identified as an oncogene in nasopharyngeal carcinoma.15 Bai et al also preliminarily uncovered the biomarker role of SLC9A3-AS1 in LC.14 However, the full-scale role of SLC9A3-AS1 in the oncogenesis and progression of NSCLC remains undefined. In this study, we revealed that SLC9A3-AS1 was upregulated in NSCLC. SLC9A3-AS1 possessed a satisfactory ability to distinguish NSCLC from LBL or HC. A high level of SLC9A3-AS1 was closely associated with poor clinicopathological features and survival outcomes in patients with NSCLC. Functionally, SLC9A3-AS1 knockdown inhibited cell proliferation, migration, and invasion of NSCLC cells. Overall, our data disclosed the diagnostic, prognostic, and oncogenic roles of SLC9A3-AS1 in NSCLC. It is relatively easy to detect SLC9A3-AS1 in blood samples by RT-qPCR, which may prove to be a promising biomarker for the clinical diagnosis of NSCLC, especially for distinguishing patients with NSCLC from those with LBC or healthy individuals. In addition, patients with overexpression of SLC9A3-AS1 may have poor prognosis, thus more comprehensive treatments should be offered after surgery for such patients.
Mounting evidence has revealed that primarily cytoplasmic located lncRNAs could function as ceRNAs for miRNAs to regulate tumorigenesis.21 In this study, subcellular fractionation assays verified that SLC9A3-AS1 was primarily distributed in the cytoplasm of NSCLC cells. Bioinformatics analysis presented that miR-760 could be a target of SLC9A3-AS1. Moreover, an inverse relationship between SLC9A3-AS1 and miR-760 was determined in NSCLC tissues. The luciferase reporter and RIP assays further confirmed the direct interaction between SLC9A3-AS1 and miR-760. Finally, the rescue experiments revealed that downregulation of miR-760 could reverse the modulatory activities of SLC9A3-AS1 knockdown on NSCLC cells. Generally, SLC9A3-AS1 functions as a ceRNA of miR-760 to regulate the malignant phenotypes of NSCLC.
Notably, miR-760 has been reported to be dysregulated in numerous human cancers.22 For example, miR-760 expression was downregulated in prostate cancer,23 gastric cancer,24 colorectal cancer,25 esophageal squamous cell carcinoma,26 and NSCLC.27 Overall, the commonly unveiled role of miR-760 is a tumor suppressor; however, in ovarian cancer, upregulated miR-760 acted as an oncogene to promote cell proliferation and migration.28 In this study, miR-760 was observed to be downregulated in NSCLC and silencing miR-760 exerted a tumor-promoting effect during NSCLC progression, which echoes with the previous findings of miR-760 in NSCLC.
In conclusion, this study demonstrated that SLC9A3-AS1 exerted oncogenic effects on NSLC cells via targeting miR-760. SLC9A3-AS1 could be a promising diagnostic and prognostic biomarker for NSCLC.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10(1):3–7. doi:10.1111/1759-7714.12916
3. Remark R, Becker C, Gomez JE, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191(4):377–390. doi:10.1164/rccm.201409-1671PP
4. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet (London, England). 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
5. Donington JS, Koo CW, Ballas MS. Novel therapies for non-small cell lung cancer. J Thorac Imaging. 2011;26(2):175–185. doi:10.1097/RTI.0b013e3182161709
6. Jones GS, Baldwin DR. Recent advances in the management of lung cancer. Clin Med. 2018;18(Suppl 2):s41–s6. doi:10.7861/clinmedicine.18-2-s41
7. Naylor EC, Desani JK, Chung PK. Targeted therapy and immunotherapy for lung cancer. Surg Oncol Clin N Am. 2016;25(3):601–609. doi:10.1016/j.soc.2016.02.011
8. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
9. Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34(2):153–180. doi:10.1080/02648725.2018.1471566
10. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
11. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–2100. doi:10.1111/cas.13642
12. Wei J, Liu X, Li T, Xing P, Zhang C, Yang J. The new horizon of liquid biopsy in sarcoma: the potential utility of circulating tumor nucleic acids. J Cancer. 2020;11(18):5293–5308. doi:10.7150/jca.42816
13. Osielska MA, Jagodziński PP. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: what do we know so far? Biomed Pharmacother. 2018;101:322–333. doi:10.1016/j.biopha.2018.02.099
14. Bai Y, Qu Y, Wu Z, et al. Absolute quantification and analysis of extracellular vesicle lncRNAs from the peripheral blood of patients with lung cancer based on multi-colour fluorescence chip-based digital PCR. Biosens Bioelectron. 2019;142:111523. doi:10.1016/j.bios.2019.111523
15. Li J, Li D, Zhang X, Li C, Zhu F. Long noncoding RNA SLC9A3‑AS1 increases E2F6 expression by sponging microRNA‑486‑5p and thus facilitates the oncogenesis of nasopharyngeal carcinoma. Oncol Rep. 2021;46(2). doi:10.3892/or.2021.8116
16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
18. Ye R, Tang R, Gan S, et al. New insights into long non-coding RNAs in non-small cell lung cancer. Biomed Pharmacother. 2020;131:110775. doi:10.1016/j.biopha.2020.110775
19. Zhao L, Zhang X, Shi Y, Teng T. LncRNA SNHG14 contributes to the progression of NSCLC through miR-206/G6PD pathway. Thorac Cancer. 2020;11(5):1202–1210. doi:10.1111/1759-7714.13374
20. Yu X, Lin Q, Liu F, Yang F, Mao J, Chen X. LncRNA TMPO-AS1 facilitates the proliferation and metastasis of NSCLC cells by up-regulating ERBB2 via sponging miR-204-3p. Int J Immunopathol Pharmacol. 2020;34:2058738420958947. doi:10.1177/2058738420958947
21. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
22. Manvati MKS, Khan J, Verma N, Dhar PK. Association of miR-760 with cancer: an overview. Gene. 2020;747:144648. doi:10.1016/j.gene.2020.144648
23. Wang S, Yang Y, Cao YD, Tang XX, Du P. Androgen downregulation of miR-760 promotes prostate cancer cell growth by regulating IL6. Asian J Androl. 2021;23(1):85–90. doi:10.4103/aja.aja_20_20
24. Liu W, Li Y, Feng S, Guan Y, Cao Y. MicroRNA-760 inhibits cell viability and migration through down-regulating BST2 in gastric cancer. J Biochem. 2020;168(2):159–170. doi:10.1093/jb/mvaa031
25. Cao L, Liu Y, Wang D, et al. MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J Exp Clin Cancer Res. 2018;37(1):83. doi:10.1186/s13046-018-0757-8
26. Yang X, Zhang C, Tie H, Luo J, Wang Y, Wu Q. miR-760 exerts an antioncogenic effect in esophageal squamous cell carcinoma by negatively driving fat metabolism via targeting c-Myc. J Cell Biochem. 2020;121(4):2950–2961. doi:10.1002/jcb.29540
27. Wang W, He B. MiR-760 inhibits the progression of non-small cell lung cancer through blocking ROS1/Ras/Raf/MEK/ERK pathway. Biosci Rep. 2020. doi:10.1042/BSR20182483
28. Liao Y, Deng Y, Liu J, et al. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol. 2016;143(3):655–663. doi:10.1016/j.ygyno.2016.09.010
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.