Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle
Authors Qiu X, Dong J, Zhao Z, Li J, Cai X
Received 27 September 2018
Accepted for publication 6 March 2019
Published 11 July 2019 Volume 2019:12 Pages 5615—5625
DOI https://doi.org/10.2147/OTT.S188933
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Xia Qiu, Jiangnan Dong, Zheng Zhao, Jun Li, Xiaoyan Cai
Department of General Surgery, Pudong New Area Gongli Hospital Affiliated to Naval Military Medical University, Shanghai 200135, People’s Republic of China
Objective: To elucidate how lncRNA 00668 (LINC00668) influences the development of breast cancer (BC).
Materials and methods: Genome-wide expression profile of BC and paracancerous tissues were downloaded from The Cancer Genome Atlas (TCGA) and BC tissues and paracancerous tissues enrolled from our hospital for analyzing the expression level of LINC00668 and its correlation with prognosis. GSEA was conducted to analyze the potential functions of LINC00668. By transfection of sh-LINC00668 in BC cells, proliferation, apoptosis, cell cycle and colony formation of BC cells were accessed. Western blot was conducted to detect protein expressions of Ki-67, CDK4, Bcl-2, p21 and genes in AKT/mTOR pathways after LINC00668 knockdown in BC cells. Finally, tumor-bearing nude mice were administrated with BC cells. We compared the proliferative rate in mice with different administrations. Immunohistochemistry was carried out to access expression levels of Ki-67, CDK4, Bcl-2 and P21 in mice.
Results: Both TCGA data and BC tissues harvested from our hospital indicated the higher expression of LINC00668 in BC tissues. LINC00668 expression was negatively correlated to prognosis of BC patients. GSEA pointed out that LINC00668 is enriched in regulations of cell cycle and apoptosis. By transfection of sh-LINC00668 in MDA-MB-231 and MDA-MB-436 cells, the proliferative and colony formation abilities of BC cells decreased. Besides, LINC00668 knockdown in BC cells induced apoptosis and arrested cell cycle. LINC00668 knockdown downregulated Ki-67, CDK4 and Bcl-2, but upregulated p21. The AKT/mTOR pathway was inhibited after LINC00668 silenced. In vivo experiments demonstrated the decreased proliferative rate in tumor-bearing mice administrated with sh-LINC00668 transfected BC cells. Consistently, immunohistochemical results showed lower positive expressions of Ki-67, CDK4 and Bcl-2, but higher positive expression of p21 in sh-LINC00668 group.
Conclusion: LINC00668 is highly expressed in BC tissues and can promote the progression of BC by inhibiting apoptosis and accelerating cell cycle progression.
Keywords: LINC00668, breast cancer, apoptosis, cell cycle
Introduction
Breast cancer (BC) is one of the most common malignancy in women,1 and its incidence rate is the highest in female tumors worldwide. Younger onset age of BC seriously threatens physical and mental health of females.2 The occurrence of BC is a complex biological process involving multiple genes, factors and pathways.3 With the rapid development of molecular biology, we found differentially expressed genes that are associated with various biological behaviors of BC through the analysis of gene expression profiles. Currently, comprehensive treatments include surgery, radiotherapy and chemotherapy for BC have been well applied. However, the low detective rate of early-stage BC limits its therapeutic efficacy. Hence, it is of great significance to elucidate the molecular mechanism of BC, so as to improve early diagnostic rate and clinical outcomes of BC.
LncRNA is a kind of RNA molecule with a length of 200 nt and it could not encode proteins.4 Recent studies have confirmed the role of lncRNAs in the occurrence and progression of diseases.5 LncRNAs participate in multiple biological processes, such as proliferation, apoptosis, differentiation, metastasis and stem cell pluripotency.5–7 The regulatory forms of lncRNAs are multifaceted, and they could regulate gene expressions at epigenetic, transcriptional and post-transcriptional levels.8 It is reported that lncRNAs are involved in the tumor development as oncogenes or tumor-suppressor genes.8,9 In particular, some certain lncRNAs have exerted their remarkable roles in BC development.10,11 For example, lncRNA H19 is highly expressed in BC tissues, which is associated with ER and PR. LncRNA H19 regulates proliferation and invasion of BC by regulating HIF-1α, P53 and E2F1.12 LncRNA MALAT1 is highly expressed in BC tissues and cell lines, which is capable of regulating BC proliferation by downregulating the expression of miR-124.13 LncRNA UCA1 promotes proliferative ability of BC cells by inhibiting protein level of P27. UCA1 knockdown markedly arrests tumor cell cycle.13 Therefore, lncRNAs are significant in the development of BC. Developing novel therapeutic target based on lncRNAs may contribute to improve the therapeutic efficacy of BC.
Cell cycle is the process by which a cell divides to two daughter cells, which is the basic feature of cellular activity.14 Since the cell cycle runs through the entire cellular life process, it is closely related to a variety of cellular biological behaviors.15 New cells are continuously produced to participate in cell proliferation. The coordination and control of cell cycle, proliferation, differentiation and apoptosis exert an important role in the regulation of tumors, immune diseases, neurodegenerative diseases and viral infections.16,17 Studies have shown the crucial functions of lncRNAs in cell cycle regulation. LncRNA GAS5 inhibits proliferative rate of gastric cancer cells by accelerating cell cycle arrest in G1 phase through P21 regulation.18 LncRNA PANDAR is highly expressed in thyroid cancer cells. Interference with PANDAR expression could suppress proliferation and cell cycle but induces apoptosis of thyroid cancer cells.19
This study elucidated the regulatory role of LINC00668 in proliferative and apoptotic rate of BC cells. Our study provides references for diagnosis and treatment of BC.
Methods
Sample collection
Patients with BC treated in our hospital from 2007 to 2017 were enrolled. None of them received any preoperative treatments. All BC tissue samples were pathologically diagnosed. Paracancerous tissues were harvested at 5 cm away from the BC tissues and confirmed without any infiltration. Tissue samples were immediately preserved in liquid nitrogen. Sample collection was obtained from the written informed consent of patients and approved by the ethics committee of Naval Military Medical University and in accordance with the 1964 Helsinki declaration and its later amendments.
Data collection
Public breast cancer susceptibility genes transcriptome database (RNASeqV2: https://wiki.nci.gov.display/TCGA/RNASeq+Version+2) and clinical data of BC samples (data version: 2018–01-28) were downloaded from http://gdac.broadinstitute.org/, including 1393 cases of BC samples and 212 cases of paracancerous tissues.
Survival analyses
BC samples were divided into two high and low expression groups using the survivalROC package with the gene expression boundary value of 3.225207. The overall survival curve was introduced using the survival package.
Gene set enrichment analysis (GSEA)
According to the expression level of LINC00668 in BC tissues, they were divided into high and low expression group. Data processing was conducted to identify the enriched pathways with the criteria of false discovery rates<0.25 and P<0.01 using GSEA 2.2.1 software.
Cell culture
BC cell lines (T47D, MCF7, MDA-MB-231and MDA-MB-436) and normal breast cell line (MCF-10A) were obtained from ATCC. MCF-10A cells were cultured in DMEM containing 10% FBS, and the others were cultured in RPMI 1640 containing 10% FBS. Cells were maintained in a 5% CO2 incubator at 37°C.
Cell transfection
Construction of sh-LINC00668, vector ligation and transformation, preparation of competent cells, lentiviral packaging and recombinant lentiviral titer determination were completed by GenePharma, Co., Ltd. BC cells were seeded in the 24-well plate with 1×105cells per well. Until cell confluence of 30–50%, 2 mL of fresh medium containing 5 ng/mL polybrene and lentiviruses (sh-NC, sh-LINC00668 1#, 2#, 3# or 4# with MOI=20) was added in each well. QRT-PCR was performed to verify the transfection efficacy 96 hrs later. Plasmid sequences were as follows: shRNA1# sense 5ʹ-CCACUCCCAUCCACUGUAGUGUAAA-3ʹ, antisense 5ʹ-UUUACACUACAGUGGAUGGGAGUGG-3ʹ; shRNA2# sense 5ʹ-GAAUCUUGGGCGGUCUGAAAUCUGA-3ʹ; antisense 5ʹ-UCAGAUUUCAGACCGCCCAAGAUUC-3ʹ; shRNA3# sense 5ʹ- GAGGCCUAUAAUGCAAAGATT-3ʹ; antisense 5ʹ-UCUUUGCAUUAUAGGCCUCTT-3ʹ. shRNA4# sense 5ʹ-GTCATATAAGGCTGTTTCGAGCC-3ʹ; antisense: 5ʹ-GGCTAAGGAGGTGTTAACCCGAAG-3ʹ.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA in 30 mg tissues or 2×105cells was extracted using TRIzol method for reverse transcription according to the instructions of PrimeScript RT reagent Kit (Takara, Tokyo, Japan). RNA concentration was detected using a spectrometer. qRT-PCR was then performed based on the instructions of SYBR Premix Ex Taq TM (Takara, Tokyo, Japan). The amplification conditions were 50°C for 2 min, 95°C for 2 min, 95°C for 15 s and 60°C for 30 s, for a total of 40 cycles. The relative gene expression was calculated using 2−△Ct method. Primers used in the study were as follows: GAPDH (forward): 5ʹ-CGGAGTCAACGGATTTGGTCGTAT-3ʹ; GAPDH (reverse): 5ʹ-AGCCTTCTCCATGGT GGTGAAGAC-3ʹ; LINC00668 (forward): 5′-GCACCTGTCATTGCAATTCC-3′; LINC00668 (reverse): 5′- CTGTGTCTAACTTAGTGGCTC-3′.
Western blot
BC cells were lysed to harvest total cellular protein, followed by determination of total protein concentration. An equal amount of protein sample was loaded onto a 10% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane. Membranes were blocked with skim milk and incubated with primary antibody (Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. At the other day, membranes were incubated with horseradish peroxidase conjugated secondary antibody for 2–3 hrs at room temperature. Finally, an image of the protein band was captured by the Tanon detection system using ECL reagent (Thermo, Waltham, MA, USA).
Immunohistochemistry
Paraffin-embedded tissues were deparaffinized, hydrated in ethyl alcohol and incubated with 0.3% H2O2 for elimination of endogenous peroxidase activity. After rupture of nuclear membrane with 0.1% Triton X-100 for 30 mins and blockage with 5% normal donkey serum, slides were incubated with primary antibody at 4°C overnight and secondary antibody at room temperature for 1 hr. Immunohistochemistry results were captured using Nikon Eclipse 80i microscope.
EdU
BC cells were resuspended for cell density adjustment at 2×105/mL. A total of 100 μL of suspension was seeded into each well of the 96-well plate. Until cell confluence, BC cells were incubated with 10 μmol/L EdU for 4 hrs, followed by staining with AdoLo and Hochest. EdU-positive cells were observed using a fluorescence microscope at the magnification of 100×.
CCK-8 assay
BC cells were resuspended for cell density adjustment at 2×105/mL. A total of 100 μL of suspension was seeded into each well of the 96-well plate. Ten microliters of CCK-8 solution (cell counting kit-8, Dojindo, Kumamoto, Japan) was added in each well after cell culture for 24, 48, 72 and 96 hrs, respectively. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, USA).
Apoptosis determination
BC cells were collected and washed with PBS twice. Subsequently, BC cells were incubated with 500 μL of Annexin V and 5 μL of PI at room temperature for 10 mins in dark. Apoptotic cells were determined using the flow cytometry.
Colony formation assay
BC cells were prepared for cell suspension and seeded in the 6-well plates at a dose of 250, 500 and 1000 cells per well. Fresh medium was replaced every 2–3 days. Fifteen days later, cells were washed with pre-cooled PBS once and stained with 400 μL of crystal violet for 10–30 mins and captured using the microscope.
Cell cycle determination
BC cells were prepared for cell suspension and washed with pre-cooled PBS for three times. Cells were suspended in the mixture containing 0.1 mL of pre-cooled PBS and 1 mL of pre-cooled 75% ethanol. Subsequently, ethanol was washed with PBS and cells were incubated with PI and RNase A at a final dose of 50 ng/ml. After 37°C water bath for 30 mins in dark, cell cycle was determined using flow cytometry.
Construction of tumor-bearing mice model
BC cells in good condition were adjusted at a dose of 1×106/mL. Healthy BALB/c nude mice with 5 or 6 weeks old were randomly assigned into sh-NC group (n=9) or sh-LINC00668 1# group (n=9). Right chest fat pad of nude mouse was sterilized with 75% ethanol and 100 μL of BC cell suspension transfected with sh-NC or sh-LINC00668 1# was administrated. Mice were observed every 3 days. The long diameter and transverse diameter of BC tumor were measured using the vernier caliper. Volume of the BC tumor was calculated: V (mm3) =0.5× long diameter (mm) × transverse diameter2 (mm2). Mice were sacrificed 3 weeks later, and tumor samples were harvested for weight recording. The staining results were independently scored by the author and a pathologist to minimize subjectivity, and the experiments were approved by the Animal ethics committee of Naval Military Medical University.
Statistical analyses
SPSS 22.0 statistical software was used for data analysis. Data were expressed as mean±SD. Measurement data and classification data were compared using the Student's t-test. P<0.05 considered the difference was statistically significant.
Results
LINC00668 was highly expressed in BC and negatively correlated with its prognosis
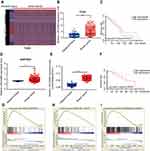
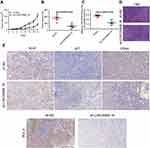
To explore relative lncRNAs in BC, we first downloaded the genome-wide expression profile data for BC and corresponding paracancerous tissues in The Cancer Genome Atlas (TCGA) (Figure 1A). Through data analyses, we found that LINC00668 is highly expressed in BC tissues compared with that of paracancerous tissues (Figure 1B). Further survival analyses revealed the negative correlation between LINC00668 expression and prognosis of BC patients (Figure 1C) in TCGA. Subsequently, we analyzed LINC00668 expression in BC tissues of GSE54002. The mRNA level of LINC00668 was markedly higher in BC tissues compared with that of paracancerous tissues in GSE54002 (Figure 1D). The differential expression of LINC00668 was identical in BC tissues and paracancerous tissues harvested from our hospital (Figure 1E). By analyzing LINC00668 expression in BC patients enrolled from our hospital, it indicated that BC patients with higher expression of LINC00668 experience a worse prognosis (Figure 1F). To further elucidate the potential biological functions of LINC00668, GSEA was utilized and the results showed that LINC00668 is mainly enriched in cell cycle and apoptosis (Figure 1G–I). The above data demonstrated that LINC00668 is closely related to the occurrence and progression of BC.
LINC00668 knockdown inhibited proliferative ability of BC cells
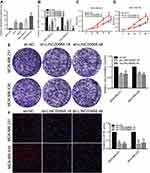
LINC00668 expression in BC cell lines (T47D, MCF7, MDA-MB-231 and MDA-MB-436) and normal breast cell line (MCF-10A) was determined by qRT-PCR. It is found that LINC00668 is highly expressed in BC cells than that of normal breast cells (Figure 2A). We selected MDA-MB-231 and MDA-MB-436 cells with relatively high expression of LINC00668 for the following experiments. Subsequently, LINC00668 interference sequences were constructed and sh-LINC00668 1# and sh-LINC00668 4# obtained the highest transfection efficacy, which markedly downregulated mRNA level of LINC00668 in MDA-MB-231 and MDA-MB-436 cells (Figure 2B). To elucidate the proliferative rate of BC cells after LINC00668 knockdown, the growth curves of BC cells transfected with sh-NC or sh-LINC00668 1# and sh-LINC00668 4# were analyzed according to the absorbance at 450 nm. CCK-8 results showed lower absorbance in BC cells transfected with sh-LINC00668 1# and sh-LINC00668 4# at 48 h, 72 h, and 96 h, respectively (P<0.05, Figure 2C and D). Besides, colony formation assay revealed that interference with LINC00668 expression inhibits the colony formation abilities of MDA-MB-231 and MDA-MB-436 cells (Figure 2E). EdU assay also suggested the decreased proliferative ability of BC cells after LINC00668 knockdown (Figure 2F). These results suggested that knocking down of LINC00668 can inhibit proliferative and colony formation abilities of BC cells.
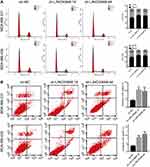
LINC00668 knockdown promoted cell cycle arrest and induced apoptosis
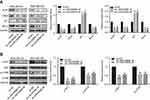
The potential influences of LINC00668 on cell cycle and apoptosis of BC cells were elucidated here. The amount of MDA-MB-231 and MDA-MB-436 cells in G0/G1 phase markedly increased, whereas cells in S phase were reduced after LINC00668 knockdown, indicating the arrested cell cycle (Figure 3A). Flow cytometry showed a large number of apoptotic cells after LINC00668 knockdown in MDA-MB-231 and MDA-MB-436 cells (Figure 3B). Western blot was then conducted to explore protein levels of relative genes in proliferation (Ki-67), cell cycle (CDK4, P21) and apoptosis (Bcl-2). LINC00668 knockdown downregulated Ki-67, CDK4 and Bcl-2, but upregulated P21 in MDA-MB-231 and MDA-MB-436 cells (Figure 4A). The AKT/mTOR pathway usually keeps in a hyperactive state in tumor cells.20 Our study found that AKT/mTOR pathway is inhibited by LINC00668 knockdown (Figure 4B). It is speculated that knocking down of LINC00668 inhibits proliferative ability of BC cells through arresting cell cycle and induced apoptosis.
LINC00668 knockdown inhibited in vivo proliferation of BC
To elucidate whether LINC00668 could exert its biological function in in vivo proliferation of BC as well, we administrated BC cell suspension transfected with sh-LINC00668 1# in nude mice. We observed that the tumorigenesis was slower in mice administrated with BC cell suspension transfected with sh-LINC00668 1# that of controls (Figure 5A). Twelve days after suspension administration, the tumor weight in mice of sh-LINC00668 1# group was remarkably lower than those of controls (Figure 5B). Besides, these mice expressed a relatively lower expression of LINC00668 in vivo (Figure 5C). H&E and immunohistochemical results showed lower positive expressions of Ki-67, CDK4 and Bcl-2, but higher positive expression of P21 in sh-LINC00668 1# group (Figure 5D and E).
Discussion
As an emerging hotspot in tumor researches, lncRNAs in BC have been well concerned. Some certain lncRNAs in BC are abnormally expressed, which may be utilized as potential therapeutic targets.21,22 For example, the expression level of lncRNA MEG3 in BC is downregulated and closely related to the prognosis of affected patients.23 LncRNA H19 is highly expressed in BC, exerting promotive functions of proliferation, invasion and drug resistance of BC cells.23,24 Hotair serves as an oncogene and is closely related to metastasis, drug resistance and poor prognosis of BC patients.25 The important roles of lncRNAs in tumors have gradually become a crucial direction in molecular biology and clinical researches. According to the data in NCBI (Gene, NR_034100.1), LINC00668 has only one transcript with 1751 bp. Current studies found that LINC00668 is upregulated in gastric cancer and can promote the proliferative capacity of gastric cancer cells.26 LINC00668 regulates VEGF signaling in oral squamous cell carcinoma by inhibiting miR-297.27 However, the specific functions of LINC00668 in BC have not been reported yet. In the present study, we found LINC00668 is highly expressed in BC tissues than that of paracancerous tissues based on the heatmap of genome-wide expression profiles of BC. Consistently, LINC00668 expression was higher in BC tissues harvested from our hospital compared with that of paracancerous tissues. Survival analysis indicated the negative correlation between LINC00668 expression and the prognosis of BC patients. Knockdown of LINC00668 in BC cells markedly arrested cell cycle, inhibited proliferative and colony formation abilities, and induced apoptosis. It is expected that LINC00668 could be used as a novel diagnostic target for BC.
It is well known that uncontrolled cell cycle would lead to excessive cell proliferation and canceration.28 The regulation of the G1 phase in cell cycle is a complex process involving multiple cell cycle regulators, which is closely related to tumor development.29 Cyclin and CDK are the core factors in cell cycle regulation. CyclinD/CDK4 and CyclinD/CDK2 are the rate-limiting steps in G1 phase,which is the major phase in initiating cell cycle for proliferation. Activation of CDK2 and CDK4 promotes transformation from G1 phase to S phase by regulating cell cycle-related proteins.29 P21 is one of the major cyclin-dependent protein kinase inhibitors. It serves as a tumor-suppressor gene that inhibits cell division or induces apoptosis by negatively regulating CDK function.30 Bearss DJ et al found the accelerated growth of BC cells in the BC model of p21WAF1/CIP deficiency mice.31 Downregulation of P21 inhibits anti-estrogen-mediated cell cycle arrest in human BC cells.22 In addition, the PI3K/AKT/mTOR signaling pathway is involved in the regulation of cell cycle.32 It is reported that PI3K/AKT pathway can upregulate the expression of Cyclin D by inhibiting the activity of GSK3-β, and meanwhile, it can also inhibit TSC2 activity and p27Kip1 expression, thus jointly promoting the progression from G1 phase to S phase.33 The PI3K/AKT/mTOR pathway regulates the progression of G0-G1-S phase by activating Cyclin D and c-Myc accompanied with the Wnt pathway.34 Our study demonstrated that LINC00668 knockdown in MDA-MB-231 and MDA-MB-436 cells markedly arrests cell cycle, inhibits proliferative and colony formation abilities through suppressing AKT/mTOR pathway.
However, some shortcomings in this study should be concerned. The prognosis of BC patients is not only closely related to the proliferation of tumor cells but also related to the enhanced migratory and invasive abilities of tumor cells. In this experiment, we were not able to evaluate the regulatory effect of LINC00668 on migratory ability of cells. Further wound healing or Transwell assay should be conducted to elucidate the migration change of BC cells induced by LINC00668. Moreover, the potential influences of LINC00668 overexpression on biological performances of BC cells are needed to be verified, so as to improve the credibility of the experimental results.
In summary, this study firstly explored the role of LINC00668 in the development of BC. LINC00668 was highly expressed in BC tissues and cell lines, and its expression was negatively correlated with the prognosis of BC patients. GSEA analysis found that LINC00668 is mainly enriched in functions such as apoptosis and cell cycle. After interfering with the expression of LINC00668, the proliferative and colony formation abilities of BC cells markedly decreased, apoptotic rate increased, and cell cycle progression was significantly blocked. LINC00668 knockdown inhibited the AKT/mTOR pathway. Consistently, in vivo experiments showed that the tumor formation ability and proliferative rate decreased after interfering with the expression of LINC00668. It is speculated that LINC00668 may act as an oncogene to promote the occurrence, development, invasion, invasion and metastasis of BC. LINC00668 is expected to become a new target for the future tumor treatment.
Conclusion
LINC00668 is highly expressed in BC tissues and can promote the progression of BC by inhibiting apoptosis and accelerating cell cycle progression.
Acknowledgment
This project was supported by the Special Project of Intergrated traditional Chinese and Western Medicine of Shanghai General Hospital (ZHYY-ZXYJHZX-201621).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Mincey BA. Genetics and the management of women at high risk for breast cancer. Oncologist. 2003;8:466–473.
2. Plevritis SK, Munoz D, Kurian AW, et al. Association of screening and treatment with breast cancer mortality by molecular subtype in us women, 2000–2012. JAMA. 2018;319:154–164. doi:10.1001/jama.2017.19130
3. Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol. 2017;14(10):595–610.
4. Frith MC, Bailey TL, Kasukawa T, et al. Discrimination of non-protein-coding transcripts from protein-coding mrna. RNA Biol. 2006;3:40–48.
5. Wang KC, Chang HY. Molecular mechanisms of long noncoding rnas. Mol Cell. 2011;43:904–914. doi:10.1016/j.molcel.2011.08.018
6. Guttman M, Donaghey J, Carey BW. Lincrnas act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295. doi:10.1038/nature10398
7. Gupta RA, Shah N, Wang KC, et al. Long non-coding rna hotair reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi:10.1038/nature08975
8. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding rnas. Cell. 2009;136:629–641. doi:10.1016/j.cell.2009.02.006
9. Zhang A, Xu M, Mo YY. Role of the lncrna-p53 regulatory network in cancer. J Mol Cell Biol. 2014;6:181–191. doi:10.1093/jmcb/mju013
10. Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. Lncrna-atb promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652. doi:10.18632/oncotarget.3457
11. Arase M, Horiguchi K, Ehata S, et al. Transforming growth factor-β-induced lncrna-smad7 inhibits apoptosis of mouse breast cancer jygmc(a) cells. Cancer Sci. 2015;105:974–982. doi:10.1111/cas.12454
12. Shore AN, Herschkowitz JI, Rosen JM. Noncoding rnas involved in mammary gland development and tumorigenesis: there’s a long way to go. Journal of Mammary Gland Biology & Neoplasia. 2012;17:43–58. doi:10.1007/s10911-012-9247-3
13. Feng T, Fang S, Wu Q, et al. Mir-124 downregulation leads to breast cancer progression via lncrna-malat1 regulation and cdk4/e2f1 signal activation. Oncotarget. 2016;7:16205–16216. doi:10.18632/oncotarget.7578
14. Agarwal ML, Agarwal A, Taylor WR, Stark GR. P53 controls both the g2/m and the g1cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8493–8497.
15. Jiang Q, Isquith J, Zipeto MA,
16. Normile D. Cell proliferation. Common control for cancer, stem cells. Science. 2002;298:1869. doi:10.1126/science.298.5600.1869
17. Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462.
18. Liu Y, Zhao J, Zhang W, et al. Lncrna gas5 enhances g1 cell cycle arrest via binding to ybx1 to regulate p21 expression in stomach cancer. Sci Rep-Uk. 2015;5:10159. doi:10.1038/srep10159
19. Li Z, Gao B, Hao S, et al. Knockdown of lncrna-pandar suppresses the proliferation, cell cycle and promotes apoptosis in thyroid cancer cells. Excli J. 2017;16:354–362. doi:10.17179/excli2017-113
20. Guertin DA, Sabatini DM. Defining the role of mtor in cancer. Cancer Cell. 2007;12:9–22. doi:10.1016/j.ccr.2007.05.008
21. Mendell JT. Targeting a long noncoding rna in breast cancer. N Engl J Med. 2016;374:2287–2289. doi:10.1056/NEJMcibr1603785
22. Prensner JR, Chinnaiyan AM. The emergence of lncrnas in cancer biology. Cancer Discov. 2011;1:391. doi:10.1158/2159-8290.CD-11-0209
23. Zhang W, Shi S, Jiang J, Li X, Lu H, Ren F. Lncrna meg3 inhibits cell epithelial-mesenchymal transition by sponging mir-421 targeting e-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312–319. doi:10.1016/j.biopha.2017.04.085
24. Matouk IJ, Raveh E, Abu-Lail R, et al. Oncofetal h19 rna promotes tumor metastasis. Biochim Biophys Acta. 2014;1843:1414–1426. doi:10.1016/j.bbamcr.2014.03.023
25. Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855.
26. Zhang E, Yin D, Han L, et al. E2f1-induced upregulation of long noncoding rna linc00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of ckis. Oncotarget. 2016;7:23212–23226. doi:10.18632/oncotarget.6745
27. Zhang CZ. Long intergenic non-coding rna 668 regulates vegfa signaling through inhibition of mir-297 in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2017;489. doi:10.1016/j.bbrc.2017.05.155
28. Evan GI. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi:10.1038/35077213
29. Massagu Eacute J. G1 cell-cycle control and cancer. Nature. 2004;432:298. doi:10.1038/nature03094
30. Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the cdk inhibitor p21 by vitamin d3 leads to the induced differentiation of the myelomonocytic cell line u937. Genes Dev. 1996;10:142–153.
31. Bearss DJ, Lee RJ, Troyer DA, Pestell RG, Windle JJ. Differential effects of p21(waf1/cip1) deficiency on mmtv-ras and mmtv-myc mammary tumor properties. Cancer Res. 2002;62:2077–2084.
32. Gao N, Zhang Z, Jiang BH, Shi X. Role of pi3k/akt/mtor signaling in the cell cycle progression of human prostate cancer. Biochem Biophys Res Commun. 2003;310:1124–1132.
33. Liang J, Slingerland JM. Multiple roles of the pi3k/pkb (akt) pathway in cell cycle progression. Cell Cycle. 2003;2:336–342. doi:10.4161/cc.2.4.433
34. Vadlakonda L, Pasupuleti M, Pallu R. Role of pi3k-akt-mtor and wnt signaling pathways in transition of g1-s phase of cell cycle in cancer cells. Front Oncol. 2013;3:85. doi:10.3389/fonc.2013.00085
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.