Back to Journals » Cancer Management and Research » Volume 12
LncRNA LEF1-AS1 Promotes Ovarian Cancer Development Through Interacting with miR-1285-3p
Received 17 August 2019
Accepted for publication 30 November 2019
Published 30 January 2020 Volume 2020:12 Pages 687—694
DOI https://doi.org/10.2147/CMAR.S227652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yanan Zhang, Fang Ruan
Department of Obstetrics and Gynecology, Affiliated Hospital of Jining Medical College, Jining 272000, People’s Republic of China
Correspondence: Fang Ruan
Department of Obstetrics and Gynecology, Affiliated Hospital of Jining Medical College, 89 Guhuai Road, Jining, Shandong 272000, People’s Republic of China
Email [email protected]
Background: Growing evidence indicates that long noncoding RNA (lncRNA) is a group of important regulator in cancer development. However, the correlation between lncRNA and ovarian cancer remains elusive. Here, we aimed to investigate the roles of LEF1-AS1 in ovarian cancer progression.
Methods: LEF1-AS1 expression was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Survival rate was analyzed by Kaplan-Meier method. Cell Counting Kit-8 (CCK8) and colony formation assays were used for proliferation analysis. Transwell assay was utilized for analyses of migration and invasion. Luciferase reporter assay was performed to test the interaction between LEF1-AS1 and miR-1285-3p.
Results: We showed that LEF1-AS1 expression was upregulated in ovarian cancer tissues compared with normal tissues. Besides, LEF1-AS1 level was positively correlated with lymph node metastasis and advanced stage. Enhanced expression of LEF1-AS1 may predict a poor prognosis. Moreover, LEF1-AS1 knockdown suppressed ovarian cancer cell proliferation, migration and invasion. Mechanistically, LEF1-AS1 exerted its oncogenic functions through interacting with miR-1285-3p to inhibit miRNA activity. Rescue assay validated that miR-1285-3p inhibitors abrogated LEF1-AS1-silencer-caused suppression of ovarian cancer progression.
Conclusion: Our study revealed that LEF1-AS1 acts as a vital regulation in ovarian cancer progression.
Keywords: ovarian cancer, LEF1-AS1, miR-1285-3p, progression
Introduction
Ovarian cancer (OC) is one of the most prevalent gynecological tumors and causes growing numbers of deaths in women around the world.1 Traditional methods for ovarian cancer treatment, including surgery, radiotherapy and chemotherapy, have improved patients lifespan.2 However, the five-year survival rate of ovarian cancer patients still remains lower than 35% because of frequent metastasis.3,4 Moreover, most patients were diagnosed with ovarian cancer at the advanced stage.5 Thus, it is important to elucidate the molecular mechanism of ovarian cancer development. And it is urgently required to find effective therapeutic targets.
Long noncoding RNAs (lncRNAs) are usually aberrantly expressed in tumor tissues, including ovarian cancer.6 Accumulating studies indicate that lncRNAs play critical functions by promoting or inhibiting tumor progression.7 Moreover, many lncRNAs are identified as potential biomarkers for tumor diagnosis and prognosis.8 And several reports indicate that lncRNAs may be possible therapeutic targets for cancer intervention.9 For example, lncRNA CADM1-AS1 is reported to be a novel indicator for gastric cancer prognosis.10 LncRNA GLCC1 regulates glucose metabolism and initiates colorectal cancer development via enhancing c-Myc stability.11 LINC00668 regulates breast cancer cell proliferation and survival to promote tumorigenesis.12 In addition, lncRNA FLJ33360 contributes to ovarian cancer development via interacting with miR-30b-3p.13 Hence, it is necessary to explore the detailed mechanism of lncRNA in the regulation of ovarian cancer progression.
Previously, lncRNA LEF1-AS1 was shown to promote progression of glioblastoma, prostate cancer, lung cancer and oral squamous cell carcinoma.14–17 Nevertheless, whether LEF1-AS1 participates in ovarian cancer remains unknown. Here, we identified that LEF1-AS1 expression was upregulated in ovarian cancer tissues and maybe a prognostic biomarker. Moreover, loss of LEF1-AS1 led to impaired growth and metastasis of ovarian cancer cells. We showed that LEF1-AS1 interacted with miR-1285-3p to inhibit its expression, inducing ovarian cancer progression. Our work highlights the importance of LEF1-AS1 in ovarian cancer.
Materials and Methods
Tissue Samples
Sixty-two-ovarian cancer tissues (metastasis: 28; non-metastasis: 34; I-II: 35; III-IV: 27) and their corresponding adjacent normal tissues were collected from Affiliated Hospital of Jining Medical College. None of them received chemotherapy or radiotherapy prior to surgery. Tissues were stored in the liquid nitrogen. This study was approved by the Ethics Committee of Affiliated Hospital of Jining Medical College and written informed consents were obtained from each patient.
Cell Lines and Transfection
All ovarian cancer cell lines and normal ovarian epithelial cell line IOSE80 were obtained from American Type Culture Collection (ATCC). Cells were cultured in Dulbecco’s modified Eagle’s medium RPMI-1640 (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (FBS). The small interfering RNA (siRNA) targeting LEF1-AS1 (5ʹ-CCUGGGUGGAUAUGGUAAUTT-3ʹ) and control siRNA (5ʹ-UUCUCCGAACGUGUCACGUTT’-3) were from Guangzhou RiboBio Co., Ltd. (Guangzhou, Guangdong, China). Cell transfection (100-nM siRNA) was performed using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA). After 48 h, the silencing efficiency was determined by qRT-PCR.
qRT-PCR
Total RNA was isolated from cancer tissues or cell lines using TRIzol (Invitrogen). Then 1μg RNA was transcribed into complementary DNA (cDNA) using PrimeScript RT reagent Kit (Takara, Kyoto, Japan), followed by qPCR analysis using the SYBR Green qPCR (Takara, Kyoto, Japan). Relative expression was normalized to U6 or GAPDH and calculated based on the 2−ΔΔCt method.18 Primer sequences were as follows: LEF1-AS1 (Forward: 5ʹ-TTTGTGTGGCCTGGACTCTC-3ʹ and Reverse: 5ʹ-AACCCCTGGGACACAAACTG-3ʹ) and GAPDH (Forward: 5ʹ-ACCCAGAAGACTGTGGATGG-3ʹ and reverse: 5ʹ-TCTAGACGGCAGGTCAGGTC-3ʹ).
CCK8 Assay
Cells (2000 cells per well) were plated into the 96-well plates and incubated for indicated days. Then CCK8 solution (Dojindo Laboratories, Kumamoto, Japan) was added and incubated for 4 h. Then the absorbance at 450 nm was measured using a microplate reader (Becton, Dickinson and Company, Franklin Lakes, NJ).
Colony Formation Assay
Five hundred cells were seeded into the 6-well plates and cultured for 14 days. Then cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet solution for 15 min. Colony numbers were finally counted.
Transwell Assay
Migration and invasion were measured using Transwell assay as previously reported.19
RNA-Pull Down
After transfection of 48 h with 50 nM biotin-labeled WT or Mut miR-1285-3p, cells were lyzed using specific lysis buffer (Ambion, Austin, TX) and the supernatant were collected by centrifugation.6 Then supernatant was incubated with M-280 streptavidin magnetic beads (Sigma). The beads were washed, and the precipitated RNA was eluted and purified using TRIzol, followed by qRT-PCR analysis.
RNA Immunoprecipitation (RIP)
RIP assay was performed to analyze the interaction between LEF1-AS1 and miR-1285-3p in ovarian cancer cells as previously reported.6
Luciferase Activity Assay
The interaction between LEF1-AS1 and miR-1285-3p was analyzed by using miRDB. Wild-type or mutant-type sequence of LEF1-AS1 containing miR-1285-3p binding site was constructed into the pmirGLO luciferase reporter vector (Promega). Cells were co-transfected with luciferase reporter vector and miR-1285-3p mimics (50 nM) or NC for 24 h. Then the luciferase activity was measured using a Dual-Luciferase Reporter Gene Assay Kit (Qianchen Biotechnology, Shanghai, China).
Statistical Analysis
The results were analyzed using SPSS 21.0 software (IBM Corp, Armonk, NY) and expressed as mean ± standard deviation. The t-test and one-way analysis of variance (ANOVA) were used to analyze p value. Kaplan-Maier-Plot was used to analyze survival rate. All experiments were at least performed three times. The difference was statistically significant at P < 0.05.
Results
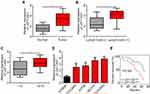
LEF1-AS1 Was Upregulated in Ovarian Cancer Tissues
To determine whether LEF1-AS1 participates in ovarian cancer development, we analyzed its expression in ovarian cancer tissues. Firstly, we collected 62 pairs of ovarian cancer tissues and adjacent normal controls. We found that LEF1-AS1 level was elevated in ovarian cancer tissues (Figure 1A). Furthermore, the expression of LEF1-AS1 was higher in ovarian cancer tissues with lymph node metastasis (Figure 1B) or advanced stages (Figure 1C). Consistently, LEF1-AS1 expression was also upregulated in ovarian cancer cell lines compared to IOSE80 cells (Figure 1D). Then, we divided the 62 patient samples into LEF1-AS1 high expression and low expression groups based on the median value of LEF1-AS1. As shown, the overall survival rate of LEF1-AS1 high expression patients were lower (Figure 1E).
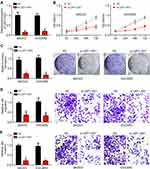
LEF1-AS1 Knockdown Inhibited Ovarian Cancer Cell Proliferation, Migration and Invasion
To explore the function of LEF1-AS1, we chose SKOV3 and OVCAR3 cells for functional experiments. We knocked down LEF1-AS1 by siRNAs (Figure 2A). CCK8 assay indicated that LEF1-AS1 knockdown inhibited the proliferation of SKOV3 and OVCAR3 cells (Figure 2B), which was further confirmed by colony formation assay (Figure 2C). To elucidate the correlation between LEF1-AS1 expression and metastasis, we carried out Transwell assay. We found that LEF1-AS1 knockdown significantly reduced the numbers of migrated and invaded SKOV3 and OVCAR3 cells (Figure 2D and E).
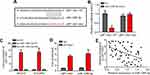
LEF1-AS1 Was the Sponge for miR-1285-3p
Then, we explored the mechanism. We searched the potential targets of LEF1-AS1 via bioinformatics analysis using the miRDB and LncBase v.2 tools. Many potential candidates were predicted, such as miR-1285-3p, miR-6860, miR-3941, miR-513c-3p and miR-612. MiR-1285-3p was identified by the both tools and had the highest predicted score. Thus we constructed the wild-type (WT) and mutant (Mut) LEF1-AS1 luciferase reporter vectors (Figure 3A). Luciferase reporter assay in SKOV3 cells indicated that LEF1-AS1-WT reporter activity was inhibited by miR-1285-3p mimics (Figure 3B). RNA-pulldown assay using biotin-labeled miR-1285-3p showed that WT bio-miR-1285-3p enriched LEF1-AS1 in cell lysates (Figure 3C). Consistently, RIP assay showed that anti-Ago2 could precipitate LEF1-AS1 and miR-1285-3p (Figure 3D). These results demonstrated that LEF1-AS1 directly interacted with miR-1285-3p. Of note, miR-1285-3p expression was negatively correlated with LEF1-AS1 in ovarian cancer tissues (Figure 3E). Thus, LEF1-AS1 interacts with miR-1285-3p and inhibits its level.
LEF1-AS1 Promoted Ovarian Cancer Progression via Inhibiting miR-1285-3p
To date, the role of miR-1285-3p in ovarian cancer remains unclear. We found that miR-1285-3p was downregulated in ovarian cancer tissues (Figure 4A). To determine whether LEF1-AS1 promotes ovarian cancer development through miR-1285-3p, we performed rescue assays. We confirmed the silencing efficiency of miR-1285-3p inhibitor (Figure 4B). Then we carried out CCK8 and Transwell assays. As shown, miR-1285-3p inhibitors could reverse the effects of LEF1-AS1 knockdown on proliferation, migration and invasion (Figure 4C–E). Moreover, only miR-1285-3p inhibition promoted proliferation, migration and invasion of SKOV3 and OVCAR3 cells (Figure 4C–E). Taken together, LEF1-AS1 promotes ovarian cancer progression via suppressing miR-1285-3p.
Discussion
Owing to rapid cell proliferation and metastasis, ovarian cancer is acknowledged as a deadly cancer.20 Of interest, several reports have demonstrated the significance of lncRNA-miRNA interaction in cancer development and progression.21 Hence, identifying novel biomarkers or therapeutic targets of lncRNA-miRNA is quite important and urgent. Here, we investigated the roles of LEF1-AS1/miR-1285-3p axis in ovarian cancer progression and demonstrated LEF1-AS1 as a new oncogene via promoting cell proliferation, migration and invasion.
Research has indicated that dysregulation of lncRNAs is correlated with ovarian cancer development.22 For example, lncRNA GAS5 upregulation suppresses the development of ovarian cancer by regulating the MAPK signaling.23 Upregulation of lncRNA HOXB-AS3 enhances ovarian cancer progression and indicates a poor prognosis.24 Our work confirmed that LEF1-AS1 was highly expressed in ovarian cancer tissues and cell lines. Previously, a study identified that LEF1-AS1 was a novel lncRNA associated with colorectal cancer metastasis and patients’ prognosis.25 Afterwards, some experiments discovered that LEF1-AS1 overexpression contributed to glioblastoma progression.14 Then, a study revealed that LEF1-AS1 promotes cell proliferation and invasion in prostate cancer by sponging miR-330-5p and upregulating LEF1 expression.15 Furthermore, it was demonstrated that LEF1-AS1 regulates growth and migration of lung cancer cells by sponging miR-544a and enhancing FOXP1 translation.26 LEF1-AS1 was also demonstrated to regulate proliferation and metastasis of oral squamous cell carcinoma and myeloid tumor.16,27 Thus, LEF1-AS1 is a very important lncRNA involved in tumorigenesis. Nevertheless, whether LEF1-AS1 regulates ovarian cancer development remains uninvestigated. In our work, we identified that LEF1-AS1 is overexpressed in ovarian cancer tissues. Furthermore, LEF1-AS1 knockdown suppressed proliferation, migration and invasion of ovarian cancer tissues. Therefore, our study convincingly shows that LEF1-AS1 exerts vital oncogenic roles in ovarian cancer.
The implication of lncRNA-miRNA axis in regulating various biological processes of tumor cells has been widely identified.28 Many reports have demonstrated the existence of lncRNA-miRNA regulatory axis in ovarian cancer, such as FLJ33360-miR-30b axis.13 A number of experiments have confirmed that LEF1-AS1 is a sponge for several miRNAs, including miR-330-5p, miR-544a and miR-489.15,17,26 LncRNA could directly interact with miRNAs to suppress their levels, consequently inducing upregulation of the miRNA downstream genes.29 For instance, LINC01116 interacts with miR-136 to promote FN1 expression in oral squamous cell carcinoma.30 In our study, via bioinformatics analysis, we identified that LEF1-AS1 was the sponge for miR-1285-3p. We demonstrated their direct interaction through luciferase reporter assay, RNA-pulldown and RIP assays. Moreover, we showed that miR-1285-3p expression was increased after LEF1-AS1 silencing in ovarian cancer cells.
Recently, the role of miR-1285-3p in ovarian cancer still remains unknown. Previous studies have indicated miR-1285-3p may be a tumor suppressor. For example, miR-1285-3p could target JUN to inhibit hepatocellular carcinoma progression.31 Besides, miR-1285-3p was reported to suppress the malignant behaviors in cervical cancer.32 Moreover, miR-1285-3p targets YAP1 to repress osteosarcoma growth and serves as a prognostic marker.33 In our study, we showed that miR-1285-3p inhibition could abrogate the effects of LEF1-AS1 knockdown on ovarian cancer progression. Moreover, we indicated that miR-1285-3p expression was downregulated in ovarian cancer tissues and miR-1285-3p silencing alone promoted cell proliferation, migration and invasion. Thus, miR-1285-3p is a tumor suppressor in ovarian cancer. In other cancer, miR-1285-3p has been shown to regulate several downstream signals, including YAP1 and JUN.31,33 Whether miR-1285-3p regulates similar signals in ovarian cancer requires further investigations.
In conclusion, our work provided evidence to support that LEF1-AS1/miR-1285-3p axis play vital roles in regulating ovarian cancer progression (Figure 5). And they may be novel therapeutic targets. However, our study is still at the preclinical stage. And the in-depth mechanism downstream LEF1-AS1/miR-1285-3p axis remains to be elucidated.
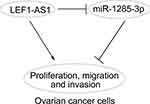 |
Figure 5 Work model. LEF1-AS1 promoted ovarian cancer progression through targeting miR-1285-3p. |
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 81502255), Medical Science and Technology Development Plans Foundation of Shandong Province (2017WS336), and Science and Technology Development Plan Foundation of Jining (No. 2014jnjc09).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yokoi A, Matsuzaki J, Yamamoto Y, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9(1):4319. doi:10.1038/s41467-018-06434-4
2. Murali R, Grisham RN, Soslow RA. The roles of pathology in targeted therapy of women with gynecologic cancers. Gynecol Oncol. 2018;148(1):213–221. doi:10.1016/j.ygyno.2017.11.020
3. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi:10.3322/caac.20138
4. Pradeep S, Kim SW, Wu SY, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26(1):77–91. doi:10.1016/j.ccr.2014.05.002
5. Moufarrij S, Dandapani M, Arthofer E, et al. Epigenetic therapy for ovarian cancer: promise and progress. Clin Epigenetics. 2019;11(1):7. doi:10.1186/s13148-018-0602-0
6. Li W, Ma S, Bai X, Pan W, Ai L, Tan W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J Cell Physiol. 2019;235:1141–1154.
7. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi:10.1158/2159-8290.CD-11-0209
8. Wang H, Fu Z, Dai C, et al. LncRNAs expression profiling in normal ovary, benign ovarian cyst and malignant epithelial ovarian cancer. Sci Rep. 2016;6:38983. doi:10.1038/srep38983
9. Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi:10.1016/j.ccr.2014.03.010
10. Shi XY, Sun YZ, Li M, Li HY. LncRNA CADM1-AS1 serves as a new prognostic biomarker for gastric cancer. Eur Rev Med Pharmacol Sci. 2019;23(3 Suppl):232–238.
11. Tang J, Yan T, Bao Y, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10(1):3499. doi:10.1038/s41467-019-11447-8
12. Qiu X, Dong J, Zhao Z, Li J, Cai X. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther. 2019;12:5615–5625. doi:10.2147/OTT.S188933
13. Yang M, Zhai Z, Guo S, Li X, Zhu Y, Wang Y. Long non-coding RNA FLJ33360 participates in ovarian cancer progression by sponging miR-30b-3p. Onco Targets Ther. 2019;12:4469–4480. doi:10.2147/OTT.S205622
14. Wang J, Liu X, Yan C, et al. LEF1-AS1, a long-noncoding RNA, promotes malignancy in glioblastoma. Onco Targets Ther. 2017;10:4251–4260. doi:10.2147/OTT.S130365
15. Liu DC, Song LL, Liang Q, Hao L, Zhang ZG, Han CH. Long noncoding RNA LEF1-AS1 silencing suppresses the initiation and development of prostate cancer by acting as a molecular sponge of miR-330-5p via LEF1 repression. J Cell Physiol. 2019;234(8):12727–12744. doi:10.1002/jcp.27893
16. Zhang C, Bao C, Zhang X, Lin X, Pan D, Chen Y. Knockdown of lncRNA LEF1-AS1 inhibited the progression of oral squamous cell carcinoma (OSCC) via Hippo signaling pathway. Cancer Biol Ther. 2019;20:1–10.
17. Yang J, Lin X, Jiang W, Wu J, Lin L. lncRNA LEF1-AS1 promotes malignancy in non-small-cell lung cancer by modulating the miR-489/SOX4 axis. DNA Cell Biol. 2019. doi:10.1089/dna.2019.4717
18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
19. Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA-340-5p. J Cell Physiol. 2019. doi:10.1002/jcp.28911
20. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. doi:10.2353/ajpath.2010.100105
21. Zhang Y, Li Y, Wang Q, et al. Identification of an lncRNAmiRNAmRNA interaction mechanism in breast cancer based on bioinformatic analysis. Mol Med Rep. 2017;16(4):5113–5120. doi:10.3892/mmr.2017.7304
22. Wang JY, Lu AQ, Chen LJ. LncRNAs in ovarian cancer. Clin Chim Acta. 2019;490:17–27. doi:10.1016/j.cca.2018.12.013
23. Long X, Song K, Hu H, et al. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38(1):345. doi:10.1186/s13046-019-1329-2
24. Zhuang XH, Liu Y, Li JL. Overexpression of long noncoding RNA HOXB-AS3 indicates an unfavorable prognosis and promotes tumorigenesis in epithelial ovarian cancer via Wnt/beta-catenin signaling pathway. Biosci Rep. 2019;39(8). doi:10.1042/BSR20190906
25. Chen Y, Yu X, Xu Y, Shen H. Identification of dysregulated lncRNAs profiling and metastasis-associated lncRNAs in colorectal cancer by genome-wide analysis. Cancer Med. 2017;6(10):2321–2330. doi:10.1002/cam4.2017.6.issue-10
26. Wang A, Zhao C, Gao Y, et al. LEF1-AS1 contributes to proliferation and invasion through regulating miR-544a/FOXP1 axis in lung cancer. Invest New Drugs. 2019. doi:10.1007/s10637-018-00721-z
27. Congrains-Castillo A, Niemann FS, Santos Duarte AS, Olalla-Saad ST. LEF1-AS1, long non-coding RNA, inhibits proliferation in myeloid malignancy. J Cell Mol Med. 2019;23(4):3021–3025. doi:10.1111/jcmm.14152
28. Panoutsopoulou K, Avgeris M, Scorilas A. miRNA and long non-coding RNA: molecular function and clinical value in breast and ovarian cancers. Expert Rev Mol Diagn. 2018;18(11):963–979. doi:10.1080/14737159.2018.1538794
29. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
30. Chen Z, Tao Q, Qiao B, Zhang L. Silencing of LINC01116 suppresses the development of oral squamous cell carcinoma by up-regulating microRNA-136 to inhibit FN1. Cancer Manag Res. 2019;11:6043–6059. doi:10.2147/CMAR.S197583
31. Liu J, Yan J, Zhou C, Ma Q, Jin Q, Yang Z. miR-1285-3p acts as a potential tumor suppressor miRNA via downregulating JUN expression in hepatocellular carcinoma. Tumour Biol. 2015;36(1):219–225. doi:10.1007/s13277-014-2622-5
32. Feng Y, Ma J, Fan H, et al. TNF-alpha-induced lncRNA LOC105374902 promotes the malignant behavior of cervical cancer cells by acting as a sponge of miR-1285-3p. Biochem Biophys Res Commun. 2019;513(1):56–63. doi:10.1016/j.bbrc.2019.03.079
33. Hu XH, Dai J, Shang HL, Zhao ZX, Hao YD. miR-1285-3p is a potential prognostic marker in human osteosarcoma and functions as a tumor suppressor by targeting YAP1. Cancer Biomark. 2019;25(1):1–10. doi:10.3233/CBM-180013
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.