Back to Journals » Cancer Management and Research » Volume 13
LncRNA HOXA-AS2 Promotes Tumor Progression by Suppressing miR-567 Expression in Oral Squamous Cell Carcinoma
Authors Chen R, Wang X, Zhou S, Zeng Z
Received 12 February 2021
Accepted for publication 13 June 2021
Published 8 July 2021 Volume 2021:13 Pages 5443—5455
DOI https://doi.org/10.2147/CMAR.S305946
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Rui Chen,1 Xi Wang,2,3 Shixian Zhou,2,4 Zongyue Zeng2
1Department of Oral and Maxillofacial Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 2Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 3Key Laboratory of Diagnostic Medicine Designated by the Ministry of Education, Department of Laboratory Medicine, Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 4Department of Pathology, Central Hospital of Jiangjin District, Chongqing, 402260, People’s Republic of China
Correspondence: Zongyue Zeng
Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, No.1 Youyi Road, Yuzhong District, Chongqing, 400016, People’s Republic of China
Tel +86 23-8895-5532
Email [email protected]
Introduction: Growing evidence suggests that long non-coding RNAs (lncRNAs), such as lncRNA HOXA-AS2, are critical regulators involved in human cancer. However, the biological functions and detailed mechanisms underlying how lncRNA HOXA-AS2 affects oral squamous cell carcinoma (OSCC) remain unexplored.
Methods: The expression of lncRNA HOXA-AS2 and miR-567 was determined in OSCC cell lines and clinical tissues by quantitative real-time PCR (qRT-PCR). Target site prediction and luciferase report assays were used to explore their potential interaction and binding sites between lncRNA HOXA-AS2 and miR-567. Overexpression or silencing expression of lncRNA HOXA-AS2 was performed to confirm that miR-567 was suppressed by lncRNA HOXA-AS2. WST-1 assay, crystal staining assay, and cell cycle analysis were used to assess the cell viability and proliferation ability. The target gene of miR-567 was predicted by Targetscan and validated by luciferase report assay as well as qRT-PCR and Western Blot. Xenograft nude mice model was done to demonstrate that lncRNA HOXA-AS2 promoted cell proliferation via targeting miR-567/CDK8 in vivo.
Results: LncRNA HOXA-AS2 was up-regulated in OSCC cells and tissues with the expression of miR-567 decreased. The tissue lncRNA HOXA-AS2 expression was found to positively correlate with the TNM stage and lymph node metastasis of OSCC patients. In terms of the mechanism, we found that lncRNA HOXA-AS2 negatively regulates miR-567 expression via a direct interaction. Functionally, overexpression of lncRNA HOXA-AS2 significantly promoted OSCC cell proliferation, while knockdown of lncRNA HOXA-AS2 significantly inhibited it. We also observed that miR-567 directly targets the 3ʹ UTR of CDK8. Moreover, silencing lncRNA HOXA-AS2 inhibited tumor growth with the expression of miR-567 increased and CDK8 decreased in vivo.
Conclusion: LncRNA HOXA-AS2 was up-regulated in OSCC, and its up-regulation correlated with poor clinical outcomes. The lncRNA also promoted OSCC cell proliferation by directly binding to miR-567, leading to an increase in CDK8 expression. The potential prognostic value of lncRNA HOXA-AS2 should be explored in future studies.
Keywords: LncRNA HOXA-AS2, MiR-567, CDK8, Oral squamous cell carcinoma, Tumor progression
Introduction
Oral squamous cell carcinoma (OSCC), the most prevalent type of head and neck cancer, is an aggressive malignant cancer, which often leads to tumor invasion and distant metastasis.1 The 5-year survival rate of OSCC patients remain about 60%, and has not shown any marked improvement in recent decades.2 The identification of specific molecular changes in a tumor can be helpful for assessing the prognosis and may be useful for targeted therapy. Therefore, there is an urgent need to identify the molecular changes present in OSCC and to explore their regulation and potential clinical applications.
With the rapid development of second-generation RNA sequencing, an increasing number of non-coding RNAs have been identified and recognized to play significant roles in tumor biogenesis and progression. Long non-coding RNA sequences (lncRNA) are large (>200 nucleotide) non-coding RNA that participate in various cellular processes through multiple mechanisms, including direct gene regulation and by functioning as competing endogenous RNA through interactions with miRNAs.3 MiRNAs are another type of non-coding RNA that down-regulate gene expression at the post-transcriptional level via binding to the 3ʹ-untranslated region (UTR) of targeted genes.4 Recently, both dysregulated miRNAs and lncRNAs were reported to be crucial mediators of gene regulating networks, forming a lncRNA-miRNA-mRNA axis, many of which are closely related to tumor growth and development.5–9
LncRNA HOXA-AS2 is up-regulated in non-small cell lung cancer and prostate cancer, and may promote tumor progression.10,11 LncRNA HOXA-AS2 was reported to regulate gene expression via a lncRNA-miRNA-mRNA axis. Cui et al12 discovered that lncRNA HOXA-AS2 promoted non-small cell lung cancer cell growth and metastasis via sponging (binding and “soaking up”) miR-520a-3p. What’s more, lncRNA HOXA-AS2 was also reported to interact with several other miRNAs, such as miR-520c-3p, miR-124-3p, and miR-145-3p.13–15 MiR-567 is located on chromosome 3q13.2 and encoded by the miR-567 gene.16 Previous studies have found that miR-567 plays significant roles in the inhibition of tumor progression and development in breast cancer,17 lung cancer,18 osteosarcoma,19 and gastric cancer.20 However, the role of lncRNA HOXA-AS2 and miR-567 in OSCC remains unclear. Therefore, we proposed the hypothesis that lncRNA HOXA-AS2 promotes tumor progression by suppressing (binding and “soaking up”) miR-567 in OSCC?
In this study, we first examined the expression of lncRNA HOXA-AS2 and miR-567 in OSCC cell lines and tumor tissues, and then determined whether its expression was associated with OSCC tumor progression in patients, and the mechanism underlying these effects.
Materials and Methods
Cell Culture and Chemicals
The OSCC cell lines, including NHOK, TSCCA, Cal-27, SCC-9, and HEK293, were purchased from the American Type Culture Collection (ATCC) in China. All cells were maintained as indicated in their instructions. Unless otherwise specified, the cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products), containing penicillin and streptomycin, as described previously.21–23 All chemicals were purchased from Sigma-Aldrich or Thermo Fisher Scientific. All of the DNA oligonucleotides were synthesized by BGI Genomics in China. The oligo sequences are listed in Supplemental Table 1.
Clinical Samples
The Committee for Ethical Review of Research Involving Human Subjects of The First Affiliated Hospital of Chongqing Medical University approved the study according to the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS). Written informed consent was provided by all patients. Human OSCC samples and adjacent tissues were collected by surgeons from the Department of Oral and Maxillofacial Surgery at The First Affiliated Hospital of Chongqing Medical University. Meanwhile, all the experiments on clinical samples were finished in accordance with the Declaration of Helsinki. The pathological diagnoses were made by three pathologists. The clinical characteristics of the 46 OSCC patients are presented in Table 1.
 |
Table 1 Correlation Between LncRNA HOXA-AS2 Expression and the Clinical Characteristics of OSCC |
Total RNA Extraction from Cells and Tissues
The total RNA from cultured cells was extracted with NucleoZOL RNA Isolation kits (Takara Bio USA, Mountain View, CA) according to the manufacturer’s instructions.22–24 The tissues were ground in an autoclaved grinding bowl with liquid nitrogen. The tissues were then treated using the total cellular RNA extraction protocol. Briefly, 20 mg of clinical tissue specimen were ground with liquid nitrogen and lysed with 500 µL NucleoZOL for 60 min at 4°C. Then, 200 µL RNase-free ddH2O was added to the lysates, and the mixture was vortexed vigorously. Subsequently, 500 µL of the supernatant was aliquoted into a new tube and precipitated with 500 µL isopropanol. After washing the resulting pellet twice with 75% ethanol, the total RNA was dissolved in 30 µL RNase-free ddH2O.
Reverse Transcription and Touchdown-qPCR (TqRCR)
To assess the expression of the lncRNA or genes of interest, reverse transcription reactions for the total RNA and TqPCR were performed as described before.22,25 All of the qPCR primers were designed and optimized using the Primer3 Plus software with the same parameters. A SYBR Green mixture (Bimake, Houston, TX) was used for the TqPCR. The reference gene was GAPDH, and the results were normalized to GAPDH using the 2−ΔΔCt method. To assess the miR expression levels, the reverse transcription reactions were carried out using miR-specific reverse primers with six nucleotides that complemented the 3ʹ end, preceded with a 44-nt artificial stem-loop sequence.24 TqPCR was also performed to quantitatively assess the miR expression levels. The results for miRs were normalized to 5S RNA by using the 2−ΔΔCt method. The SYBR Green mixture (Bimake, Houston, TX) was also used for the TqPCR according to manufacturer’s instructions.
WST-1 Cell Proliferation Assay
The WST-1 assay was performed to detect the proliferation of OSCC cells. A total of 3000 cells were pre-seeded into a 96-well plate. The absorbance was detected following the manufacturer’s protocol (Sigma-Aldrich) at 0 h, 24 h, 48 h, and 72 h. The absorbance value was used to generate the cell proliferation curve.
Cell Cycle Analysis
The TSCCA and SCC-9 cells were cultured in 35 mm dishes and treated for 48 h. After harvested, the cells were fixed with 70% ice-cold ethanol at 4 °C for overnight. The fixed cells were washed with cold PBS twice, and stained with propidium iodide (PI, 50 μg/mL) containing DNase-free RNase A (50 μg/mL) for 20 min at room temperature. Next, the cells were subjected to fluorescence-activated cell sorting (BD Biosciences, USA). The data were analyzed by ModFit software.
Plasmid Construction
The whole sequence of LncRNA HOXA-AS2 was amplified from HEK-293 genomic DNA by High-Fidelity PCR. Following digestion by HindIII and EcoRI, the fragments were cloned to generate pSEB-LncHOXA-AS2. The empty pSEB vector was used as a control (pSEB-Ctrl). To make the gene silencing expression construct, three siRNA cassettes designed using siDirect Version 2.0 were inserted into the pSEB-361-BSG vector using cassette cloning as described previously,11 resulting in pSEB-si-LncHOXA-AS2. Wild type (WT) and mutated (M1, M2) LncHOXA-AS2 were cloned into the pSEB-Gluc vector to yield pSEB-LncHOXA-AS2-WT/M1/M2 using the HindIII and EcoRI sites. The mutated LncHOXA-AS2 (M1, M2) fragments were amplified from WT LncHOXA-AS2 using site-directed mutagenesis PCR with the mutation sites in the primer. In brief, the spiked primer with a defined mutation was amplified for both side products. These products were purified and mixed at a 1:1 ratio (molecular numbers) for the 2nd step. In the second step, the full-length LncHOXA-AS2 was amplified to obtain the mutated LncHOXA-AS2 (M1, M2) fragments. Using a similar strategy, we also constructed pSEB-CDK8-WT and pSEB-CDK8-MUT.
Gene Overexpression or Silencing
Cell lines with stable overexpression or silencing of LncRNA HOXA-AS2 were established using a retrovirus packaging system as described before.11,23 Briefly, the plasmids for gene overexpression (such as pSEB-LncHOXA-AS2 or its empty control) or gene silencing (pSEB-si-LncHOXA-AS2 or its empty control) were packaged with pSEB-Ampho and then co-transfected into 293-PA cells. The culture medium was collected every 12 hours beginning on Day 2, and the media were mixed and filtered for targeted cell infection.
Western Blot
The OSCC cells were washed with pre-cold PBS twice and treated with RIPA lysis buffer for 3 h at 4°C. And then the cell lysates were centrifuged to collect the supernatants at 4°C, 12000×g for 30 min. Protein concentration was determined using protein quantitation BCA kit (QIAGEN). The proteins were separated by 4–12% PAGE gel electrophoresis and transferred to nitrocellulose membrane. Blotted membranes were blocked with 5% milk and incubated with their respective primary antibodies at 4°C overnight. After washing with PBST three times, the membranes incubated with horseradish peroxidase (HRP)-conjugated second antibody at 37°C for 1 h. The membranes were detected by using the enhanced chemiluminescence system (Bio-rad).
Luciferase Activity Assay
The previously described retrovirus packaging system23 was used to package pNRGluc-LncRNA HOXA-AS2-WT or pNRGluc-LncRNA HOXA-AS2-Mut1/2 into retroviruses. After obtained stable cell lines, we transiently transfected the cells with 1 µg pSEB-miR-567. After 48 hours, the cell culture medium was refreshed. Another 12 hours later, 100 µL cell culture medium was taken out for a Gaussia luciferase assay. The BioLux® Gaussia Luciferase Flex Assay Kit was used to detect Gaussia luciferase activity. At the same time, cells were collected for total protein qualification by the Pierce™ BCA Protein Assay Kit, which was used for data normalization. Each experiment was performed in triplicate.
Crystal Violet Staining to Assess Cell Proliferation
A total of 1.0×105 OSCC cells (TSCCA and SCC-9) with LncRNA HOXA-AS2 overexpression or silencing were cultured in 35 mm dishes for five days. For the colony forming assay, 1000 OSCC cells were grown in 35 mm dishes for 21 days. To determine the cell viability or clonal growth, the cells were stained with crystal violet staining solution (containing 4% formalin) as described previously.26
In vivo Tumor Growth Assay
The animal use and care was approved by the Committee for Ethical Review of Research of The First Affiliated Hospital of Chongqing Medical University, and all animal experiments were performed according to the Guidelines for the Ethical Review of Laboratory Animal Welfare National Standard GB/T 35,892–2018 issued by the People’s Republic of China.27 The in vivo study was also carried out in compliance with the ARRIVE guidelines 2.0.28 In brief, TSCCA or SCC-9 cells with LncRNA HOXA-AS2 silencing were harvested and re-suspended in sterile PBS, and injected subcutaneously (60 µL per injection) into the flanks of athymic BALB/c nude mice (n = 3/group, female, 4–5 week old; 2×107 cells per injection site). After four weeks, the mice were sacrificed. The tumor masses were excised, and their weight, length and width were measured. Tumor size was calculated as: volume = length × width2/2.
Statistical Analysis
All quantitative studies were carried out in triplicate. The Pearson correlation was analyzed by GraphPad Prism 6.0. A P-value < 0.05 was defined as statistically significant.
Results
The Expression of LncRNA HOXA-AS2 and miR-567 in OSCC Clinical Tissue Samples and Cell Lines
To test the hypothesis that lncRNA HOXA-AS2 promotes tumor progression by suppressing miR-567 in OSCC, we firstly evaluated the LncRNA HOXA-AS2 expression in 46 pairs of OSCC and their adjacent normal tissues by qRT-PCR. LncRNA HOXA-AS2 was expressed at a significantly higher level in OSCC tissues compared to the adjacent normal tissues (P < 0.01, Figure 1A). While analyzing the correlation between LncRNA HOXA-AS2 expression and the clinical characteristics of OSCC, we found that high expression of LncRNA HOXA-AS2 was positively related to the TNM stage and presence of lymph node metastasis (P < 0.05, Table 1). Interestingly, we found that miR-567 was down-regulated in all 46 of the paired OSCC tissues (P < 0.05; Figure 1B), and the expression of LncRNA HOXA-AS2 negatively correlated with the expression of miR-567, with a Pearson correlation of r =−0.3164 (P < 0.05, Figure 1C). Further, the expression of LncRNA HOXA-AS2 in OSCC cell lines, including TSCCA, Cal-27, and SCC-9 cells, was higher than that in a normal human oral keratinocyte cell line (NHOK) cell lines (Figure 1D); while the miR-567 expression was down-regulated in these cell lines (Figure 1E). These results suggest that lncRNA HOXA-AS2 is elevated in OSCC, and might be involved in OSCC development via miR-567.
LncRNA HOXA-AS2 Negatively Regulates miR-567 Expression by Direct Binding
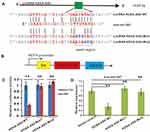
According to the biochemical basis of miRNA binding affinity proposed by David P. Bartel, 8mer, 7mer-m8, 7mer-A1, and 6mer sites to miRNA are the most effectively binding sites.29 Through sequencing alignment, we found that lncRNA HOXA-AS2 contained an 8mer potential binding site of miR-567 in the seed region with the highest binding affinity (Figure 2A). To validate this prediction, we built two constructs with mutations in the miR-567 seed region binding sites into the Gaussia Luciferase Reporter (Figure 2B). The luciferase reporter assay showed that the relative luciferase activities in lncRNA HOXA-AS2-WT and miR-567 co-transfected OSCC cells were greatly inhibited compared to those in the lncRNA HOXA-AS2-WT and miR-NC co-transfected group. In contrast, the luciferase activities were not affected in lncRNA HOXA-AS2-MUT1 or lncRNA HOXA-AS2-MUT2 and miR-567 or miR-NC co-transfected OSCC cells (Figure 2C). Further, the expression of miR-567 was down-regulated in OSCC cells with lncRNA HOXA-AS2-WT overexpression, however, the cells with overexpression of lncRNA HOXA-AS2-MUT1 or lncRNA HOXA-AS2-MUT2 did not show any change the expression level of miR-567 (Figure 2D). These results indicate that lncRNA HOXA-AS2 can regulate the expression of miR-567 via a direct interaction.
Cell Proliferation Was Promoted in lncRNA HOXA-AS2 Overexpressing OSCC Cell Lines
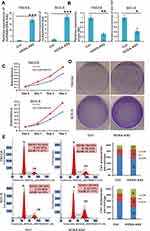
To validate the above hypothesis, we constructed TSCCA and SCC-9 cells with stable lncRNA HOXA-AS2 expression by overexpressing pSEB-lncRNA-HOXA-AS2. Compared with the vector control, miR-567 was down regulated in both TSCCA and SCC-9 cells with lncRNA HOXA-AS2 overexpression (Figure 3A and B). To better observe the proliferation of cells, we planted 105 OSCC cells (TSCCA and SCC-9) with or without lncRNA HOXA-AS2 overexpression in 35 mm dishes. Five days later, the cells were stained with crystal violet staining buffer. The results showed that the cells with lncRNA HOXA-AS2 overexpression were growing faster compared to the control cells (Figure 3D). The WST-1 results as well as the cell cycle analysis were consistent with the crystal violet staining (Figure 3C, and E). These data indicated that the overexpression of lncRNA HOXA-AS2 promoted OSCC proliferation by G0/G1 exit, and this was likely mediated by its negative regulation of miR-567.
The Proliferation of OSCC Cells Was Suppressed by si-LncRNA HOXA-AS2 Transfection
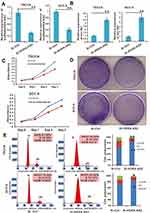
To confirm the effects of lncRNA HOXA-AS2 in OSCC cells, we silenced lncRNA HOXA-AS2 expression using the siRNA retrovirus expression system described in the Methods. The TSCCA-si-LncRNA HOXA-AS2 and SCC-9-si-LncRNA HOXA-AS2 cells lines were confirmed to have stable expression of siRNA targeting lncRNA HOXA-AS2. In these cells, lncRNA HOXA-AS2 was silenced, while miR-567 was up-regulated (Figure 4A and B). WST-1 cell viability assay, crystal violet staining experiments, and cell cycle analysis showed that silencing LncRNA HOXA-AS2 reduced the viability of both cells lines (Figure 4C–E). These data indicated that silencing LncRNA HOXA-AS2 inhibited OSCC proliferation, which was likely mediated via the negative regulation of miR-567.
CDK8 is a Downstream Target of miR-567, and is Involved in the Regulation of OSCC Cell Proliferation
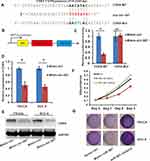
Targetscan was used to predict target genes of miR-567. It is reported that miR-567 regulates several cell proliferation related genes, such as c-Myc,20 CDK8,18 and FGF5.19 We first tested whether these genes were miR-567’s target genes in OSCC cells. And we found that only CDK8 will be down regulated in OSCC cells significantly when miR-567 was overexpressed (Supplement Figure1). MiR-567 was selected for further study. By online analysis, we found that miR-567 contained a putative binding site for CDK8 in its 3ʹUTR (Figure 5A). A luciferase reporter assay was performed to verify this prediction, and the data showed that the luciferase activities in CDK8-WT and miR-567 co-transfected SCC-9 and TSCCA cells were greatly inhibited compared to CDK8-WT and miR-NC co-transfected cells, while the luciferase activities were not affected in CDK8-MUT and miR-567/miR-NC co-transfected SCC-9 and TSCCA cells (Figure 5B and C). Subsequently, we transfected mimic miR-567, mimic control or blank control into SCC-9 and TSCCA cells and examined the expression of CDK8 at both the mRNA and protein levels. The mimic miR-567 significantly suppressed CDK8 mRNA and protein expression (Figure 5D and E). The crystal violet staining experiments and WST-1 proliferation assay showed that the miR-567 inhibitor significantly increased CDK8 expression, while the miR-567-mimic suppressed the expression of CDK8, compared to the corresponding controls (Figure 5F and G). These data revealed that CDK8 is a downstream target of miR-567 in OSCC.
Silencing lncRNA HOXA-AS2 Inhibited OSCC Tumor Growth by Releasing miR-567 to Suppress CDK8 Expression
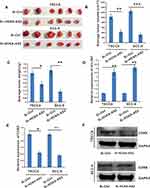
The above experiments demonstrated that the overexpression or silencing of lncRNA HOXA-AS2 increased or decreased (respectively) the proliferation of OSCC cells in vitro. We next examined whether lncRNA HOXA-AS2 influenced OSCC cell tumor growth in vivo. We subcutaneously injected TSCCA-si-LncHOXA-AS2, SCC-9-si-LncHOXA-AS2 or vector control cells into nude mice and monitored the tumor growth daily. After the mice were sacrificed at the end of the experiment, the tumors were extracted, weighed and measured. The results showed that tumors grew more slowly in the si-LncHOXA-AS2 group than in the si-ctrl group (Figure 6A–C). Additionally, it was observed that miR-567 was up-regulated in the si-LncHOXA-AS2 group (Figure 6D), while the expression of CDK8 was down-regulated in the si-LncHOXA-AS2 group (Figure 6E and F). These in vivo results strongly supported our hypothesis that si-LncHOXA-AS2 inhibits OSCC tumor growth by releasing miR-567 to suppress CDK8 expression.
Discussion
Growing evidence has shown that lncRNAs play pivotal roles in regulating gene transcription, however, the mechanisms are complex and remain largely unclear.13–15,30 Recently, there have several studies that indicated that lncRNA HOXA11-AS was up-regulated in OSCC,31 but the detailed functions and the mechanisms regulating the effects of lncRNA HOXA-AS2 in OSCC were unclear. In the present study, we investigated the involvement of lncRNA HOXA-AS2 in OSCC, and showed that lncRNA HOXA-AS2 promoted OSCC cell proliferation by sponging miR-567/CDK8. We also demonstrated that there appears to be a lncRNA HOXA-AS2-miR-567-CDK8 cell proliferation regulation network, which contributes to the development and progression of OSCC.
Our data, consistent across several OSCC cell lines, showed that lncRNA HOXA-AS2 was highly expressed in OSCC cells, and high expression of LncRNA HOXA-AS2 positively correlated with cell proliferation. In our clinical samples, lncRNA HOXA-AS2 was highly expressed in 46 OSCC patients, and the lncRNA HOXA-AS2 expression negatively correlated with the miR-567 expression, with a Pearson correlation r of −0.3164 (P <0.05). Based on these findings, we hypothesized that lncRNA HOXA-AS2 promoted OSCC cell proliferation by binding to miR-567 and acting as a miRNA sponge. To test this hypothesis, we first overexpressed lncRNA HOXA-AS2 in TSCCA and SCC-9 cells using a retrovirus packaging system. We found that overexpression of lncRNA HOXA-AS2 decreased miR-567 expression and increased the expression of its target gene, CDK8. These findings were associated with an increase in the proliferation of these cells. When we silenced lncRNA HOXA-AS2, the opposite was observed.
In addition, to explore the mechanism underlying these effects, a bioinformatic analysis was performed, and the data suggested that there was a potential miR-567 seed region binding site on lncRNA HOXA-AS2. To validate this finding, the predicted miR-567 binding site was mutated, and the mutant construct was inserted into the Gaussia Luciferase Reporter expression system. In accordance to our prediction, the mutant lncRNA HOXA-AS2 failed to inhibit miR-567 expression, and did not promote cell proliferation. Finally, based on these in vitro observations, we generated a xenograft OSCC tumor model in nude mice. Our in vivo results showed that the tumors grew slowly in the LncRNA HOXA-AS2 silenced group, which was in accordance with our in vitro data.
In summary, our current findings demonstrated that LncRNA HOXA-AS2 was highly expressed in OSCC, correlated with poorer clinical outcomes, and promoted OSCC cell proliferation via sponging miR-567. Therefore, LncRNA HOXA-AS2 appears to participate in OSCC development and progression, and may serve as a prognostic biomarker or as a potential therapeutic target.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author (Zongyue Zeng) upon reasonable request.
Ethics Approval and Consent to Participate
The animal use and care was approved by the Committee for Ethical Review of Research of The First Affiliated Hospital of Chongqing Medical University and all animal studies were performed according to the guidelines for the ethical review of laboratory animal welfare National Standard GB/T 35,892-2018 issued by the People’s Republic of China. The in vivo animal study was also carried out in compliance with the ARRIVE guidelines 2.0. The ethical consent for patient participation was approved by the Committee for Ethical Review of Research Involving Human Subjects of The First Affiliated Hospital of Chongqing Medical University according to the International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS). Written informed consent was provided by all of the patients who participated in the study. Meanwhile, all the experiments on the clinical samples were done in accordance with the Declaration of Helsinki.
Acknowledgments
We are grateful to Dr. Liuyang Zhao from The Chinese University of Hong Kong for technical support.
Funding
The reported work was supported by the Postdoctoral Program of the Natural Science Foundation of Chongqing, China (cstc2019jcyj-bsh0006 to ZZ).
Disclosure
The authors declare that they do not have any conflicts of interest.
References
1. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–316. doi:10.1016/j.oraloncology.2008.06.002
2. Hoffman HT, Porter K, Karnell LH, et al. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116(S111):1–13. doi:10.1097/01.mlg.0000236095.97947.26
3. Peng W-X, Koirala P, Mo -Y-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.184
4. Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer: figure 1. Cancer Res. 2005;65(9):3509–3512. doi:10.1158/0008-5472.CAN-05-0298
5. Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35(9):459–470. doi:10.1089/dna.2015.3187
6. Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer and Metastasis Reviews. 2009;28(3–4):369–378. doi:10.1007/s10555-009-9188-5
7. Wu K, Jiang Y, Zhou W, et al. Long noncoding RNA RC3H2 facilitates cell proliferation and invasion by targeting MicroRNA-101-3p/EZH2 axis in OSCC. Mol Ther Nucleic Acids. 2020;20:97–110. doi:10.1016/j.omtn.2020.02.006
8. Cao W, Liu JN, Liu Z, et al. A three-lncRNA signature derived from the atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi:10.1016/j.oraloncology.2016.12.017
9. Liu Z, Zhou W, Lin C, et al. Dysregulation of FOXD2-AS1 promotes cell proliferation and migration and predicts poor prognosis in oral squamous cell carcinoma: a study based on TCGA data. Aging. 2020;13:2379–2396. doi:10.18632/aging.202268
10. Cui TJ, Lin GS, Dai YM, et al. LncRNA HOXA-AS2 regulates microRNA-216a-5p to promote malignant progression of non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23:264–273. doi:10.26355/eurrev_201908_18656
11. Wang X, Yuan C, Huang B, et al. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of Short Interfering RNAs (siRNAs) in mammalian cells. ACS Synth Biol. 2019;8:2092–2105. doi:10.1021/acssynbio.9b00203
12. Liu Y, Lin X, Zhou S, Zhang P, Shao G, Yang Z. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci Rep. 2019;39.
13. Fang Y, Wang J, Wu F, Song Y, Zhao S, Zhang Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget. 2017;8:46090–46103. doi:10.18632/oncotarget.17552
14. Shi YB, Liu SL, Mou XR, et al. Long noncoding RNA HOXA-AS2 acts as an oncogene by targeting miR-145-3p in human non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2020;24:1243–1249. doi:10.26355/eurrev_202002_20177
15. Wang L, Wang L, Zhang X. Knockdown of lncRNA HOXA-AS2 inhibits viability, migration and invasion of osteosarcoma cells by miR-124-3p/E2F3. Onco Targets Ther. 2019;12:10851–10861. doi:10.2147/OTT.S220072
16. El-Murr N, Abidi Z, Wanherdrick K, et al. MiRNA genes constitute new targets for microsatellite instability in colorectal cancer. PLoS One. 2012;7:e31862. doi:10.1371/journal.pone.0031862
17. Bertoli G, Cava C, Diceglie C, et al. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res Treat. 2017;161:605–616. doi:10.1007/s10549-016-4079-2
18. Elkady MA, Doghish AS, Elshafei A, Elshafey MM. MicroRNA-567 inhibits cell proliferation and induces cell apoptosis in A549 NSCLC cells by regulating cyclin-dependent kinase 8. Saudi J Biol Sci. 2021;28:2581–2590. doi:10.1016/j.sjbs.2021.02.001
19. Liu D, Zhang C, Li X, Zhang H, Pang Q, Wan A. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI J. 2018;17:102–112. doi:10.17179/excli2017-932
20. Zhang F, Li K, Yao X, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine. 2019;44:311–321. doi:10.1016/j.ebiom.2019.05.003
21. Yan S, Zhang R, Wu K, et al. Characterization of the essential role of bone morphogenetic protein 9 (BMP9) in osteogenic differentiation of mesenchymal stem cells (MSCs) through RNA interference. Genes Dis. 2018;5:172–184. doi:10.1016/j.gendis.2018.04.006
22. Zeng Z, Huang B, Wang X, et al. A reverse transcriptase-mediated ribosomal RNA depletion (RTR2D) strategy for the cost-effective construction of RNA sequencing libraries. J Adv Res. 2020;24:239–250. doi:10.1016/j.jare.2019.12.005
23. Zeng Z, Huang B, Huang S, et al. The development of a sensitive fluorescent protein-based transcript reporter for high throughput screening of negative modulators of lncRNAs. Genes Dis. 2018;5:62–74. doi:10.1016/j.gendis.2018.02.001
24. Fan J, Feng Y, Zhang R, et al. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–437. doi:10.1038/s41417-019-0113-y
25. Zhang Q, Wang J, Deng F, et al. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR green qPCR. PLoS One. 2015;10:e0132666.
26. Shu Y, Wu K, Zeng Z, et al. System to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol Ther Nucleic Acids. 2018;13:556–567. doi:10.1016/j.omtn.2018.09.025
27. MacArthur Clark JA, Sun D. Guidelines for the ethical review of laboratory animal welfare People’s Republic of China National Standard GB/T 35892-2018 [Issued 6 February 2018 Effective from 1 September 2018]. Anim Model Exp Med. 2020;3:103–113. doi:10.1002/ame2.12111
28. Percie Du Sert N, Ahluwalia A, Alam S, et al. Reporting animal research: explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020;18:e3000411.
29. McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366(6472):366. doi:10.1126/science.aav1741
30. Yu R, Yao J, Ren Y. A novel circRNA, circNUP98, a potential biomarker, acted as an oncogene via the miR-567/ PRDX3 axis in renal cell carcinoma. J Cell Mol Med. 2020;24(17):10177–10188. doi:10.1111/jcmm.15629
31. Niu X, Yang B, Liu F, Fang Q. LncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomed Pharmacother. 2020;121:109623. doi:10.1016/j.biopha.2019.109623
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.