Back to Journals » OncoTargets and Therapy » Volume 11
lncRNA FBXL19-AS1 regulates osteosarcoma cell proliferation, migration and invasion by sponging miR-346
Authors Pan RS, He ZX, Ruan WY, Li S, Chen H, Chen ZY, Liu F, Tian XB, Nie YJ
Received 28 December 2017
Accepted for publication 17 July 2018
Published 29 November 2018 Volume 2018:11 Pages 8409—8420
DOI https://doi.org/10.2147/OTT.S160963
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Runsang Pan,1,2 Zhixu He,2 Wanyuan Ruan,2 Sun Li,1,3 Hui Chen,1,3 Zhiyu Chen,1,3 Fang Liu,1 Xiaobin Tian,1,3 Yingjie Nie1,3
1Department of Clinical Medicine, Guizhou University, Guiyang, Guizhou Province, China; 2Department of Orthopedics, Guiyang Children’s Hospital, Guiyang, Guizhou Province, China; 3Department of Orthopedics, Guizhou Provincial People’s Hospital, Guiyang, Guizhou Province, China
Background: It was recently reported that lncRNA FBXL19 antisense RNA 1 (FBXL19-AS1) is a novel tumor-promoting RNA that contributes to tumor progression by sponging miRNAs. However, the expression and function of FBXL19-AS1 in osteosarcoma (OS) have not been investigated.
Methods: Cell proliferation was assessed by the CCK-8 and colony formation assays, while cell migration and invasion were assessed using wound healing and transwell invasion assays, respectively. Quantitative reverse transcriptase PCR and immunofluorescence were used to detect the level and subcellular localization of FBXL19-AS1 expression. Interactions between miRNAs and FBXL19-AS1 were determined using luciferase reporter assays. Finally, in vivo experiments were performed to assess tumor formation.
Results: We first showed that lncRNA FBXL19-AS1 was upregulated in OS tissues and cell lines. In vitro experiments showed that FBXL19-AS1 promoted OS cell proliferation, migration, and invasion. Inhibiting miR-346 led to a significant upregulation of FBXL19-AS1, suggesting FBXL19-AS1 was negatively regulated by miR-346, which was further confirmed by the inverse correlation between FBXL19-AS1 and miR-346 expression in OS patient specimens. Furthermore, we proved that miR-346 could directly target FBXL19-AS1 through luciferase assays, suggesting FBXL19-AS1 could sponge miR-346. Additionally, inhibiting miR-346 blocked the effects of silencing FBXL19-AS1 on proliferation, migration, and invasion. Moreover, inhibiting FBXL19-AS1 significantly promoted the malignancy of MG63 and 143B cells in vivo.
Conclusion: We validated FBXL19-AS1 as a novel oncogenic lncRNA and demonstrated the molecular mechanism through which it promotes OS progression. This work advances our understanding of the clinical significance of this RNA species.
Keywords: FBXL19-AS1, miR-346, proliferation, competing endogenous RNA, osteosarcoma, lncRNA, miRNA, ceRNA, cancer
Introduction
Osteosarcoma (OS) is the most common primary malignant bone cancer in children and adolescents. Although great improvements in therapeutic strategies including radiotherapy, adjuvant chemotherapy, and surgery (wider tumor excision areas) have been achieved, the overall prognosis remains poor for most patients with recurrent or metastatic OS. Thus, there is a need to identify the molecular mechanisms underlying OS tumorigenesis and progression, to discover new specific biomarkers and therapeutic targets.
lncRNAs are a novel class of RNA transcripts of >200 nucleotides that lack protein coding potential.1 In recent years, accumulating evidence has demonstrated that lncRNAs are dysregulated in many disease states, particularly in tumors. In these diseases, lncRNAs are thought to play critical roles in the regulation of various pathophysiological processes, such as cell proliferation, apoptosis, necrosis, and autophagy.2 Recently, several lncRNAs have been reported to be involved in OS progression.3 Some classical lncRNAs previously found in various cancers were also shown to be upregulated in OS, where they promote OS progression and correlate with poor prognoses.4 However, the specific lncRNAs involved in OS pathogenesis and progression have not been clearly identified. miRNAs and lncRNAs constitute the majority of all ncRNAs.5 miRNAs are evolutionarily conserved single-stranded RNAs of 21–24 nucleotides that are involved in numerous biological processes. miRNAs play critical roles in mRNA post-transcriptional regulation by targeting the 3′ untranslated regions (UTRs) of mRNAs with their seed sequences (2–7 nucleotides in the 5′ end), resulting in mRNA degradation or translation inhibition.6
In this study, we focused on the function and regulatory mechanism of a novel lncRNA in OS. In the previous study, we showed that lncRNA FBXL19 antisense RNA 1 (FBXL19-AS1) was overexpressed in OS tumor tissues relative to adjacent normal tissues by PCR; thus, lncRNA FBXL19-AS1 might be an important molecule in OS. However, the roles of lncRNA FBXL19-AS1 in OS progression remain undefined. Based on our previous results, we further determined the role of lncRNA FBXL19-AS1 in the proliferation, migration, and invasion of OS cells in vitro and in vivo. We also found that lncRNA FBXL19-AS1 could act as a ceRNA sponge for miR-346 to further promote OS progression. Thus, lncRNA FBXL19-AS1 could be a therapeutic target for OS.
Materials and methods
Patients and cell lines
OS samples were obtained from the Department of Orthopedic Surgery, Guizhou Provincial People’s Hospital (Guiyang, China). All samples were snap frozen in liquid nitrogen immediately after resection and stored at −80°C until use. The human OS cell lines MG63, U2OS, SAOS2, HOS, 143B, and the normal osteoblast cell line hFOB1.19 were obtained from ATCC (American Type Culture Collection; Manassas, VA, USA). All cell lines were cultured at 37°C in a humidified 5% CO2 environment in DMEM or RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific).
Cell fractionation
NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) were used to prepare cytoplasmic and nuclear extracts from OS cells. RNAs extracted from each fraction were subjected to quantitative reverse transcriptase PCR (qRT-PCR) analysis to demonstrate the levels of nuclear control transcript (MALAT1), cytoplasmic control transcript (GAPDH), lncRNA FBXL19-AS1, and miR-346.
qRT-PCR analysis
Total RNA was extracted from tissues or cultured cells with TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. Reverse transcription was performed with PrimeScript RT Reagent Kit (Takara, Dalian, China). qRT-PCR was performed with the SYBR Prime Script RT-PCR Kit (Takara) according to the manufacturer’s instructions. Target gene expression was calculated with the 2−ΔΔCt method, which was normalized to GAPDH. All assays were performed in triplicate.
Colony formation assay
The osteosarcoma cells were plated into 6 cm plates (1×103 cells/plate) and incubated in DMEM or RPMI 1640 supplemented with 10% FBS at 37°C. After 2 weeks, cells were washed with PBS, fixed in methanol for 30 minutes, stained with 1% crystal violet dye, and the number of colonies were counted. All assays were performed in triplicate.
Xenograft transplantation
Female nude (BALB/c) mice (4-week-old) were purchased from Beijing Huayi Kang Company (Beijing, China) and divided into 2 groups according to a randomization method. MG63 cells (1×106) stably expressing sh-FBXL19-AS1 or a non-coding hairpin (sh-NC) were subcutaneously inoculated into the right side of the posterior flank of the mice. Tumor growth was examined at the indicated time points, and tumor volumes were measured. After 5 weeks, the mice were sacrificed, and tumors were removed and weighed.
Cell migration and invasion assay
Cellular invasion was measured using transwell chambers (8 μm pores; Corning Inc., Corning, NY, USA). Cells growing in the log phase were trypsinized, resuspended in serum-free medium, and seeded into chambers. The chambers were then inserted into transwell apparatus. The chambers were coated with Matrigel (BD Biosciences, San Jose, CA, USA) when cell invasion assays were performed. Cells were placed into the upper chamber in serum-free media 48 hours after transfection. Media containing 10% FBS were added into the lower chamber. Following a 36-hour incubation, cells in the upper chamber were wiped off, while cells that migrated to the lower membrane were fixed in methanol, stained with 0.1% crystal violet, and counted under a microscope. Three independent experiments were performed.
Wound healing assays
Cell migration was measured by wound healing assays. Briefly, 2×105 cells with or without transfection were plated into 12-well plates and incubated in DMEM or RPMI 1640 with 10% FBS at 37°C. After reaching 100% confluence, the monolayers were wounded by scraping with a 200 μL tip, followed by 3 washes with serum-free medium and incubation in serum-containing medium. Wounds were observed at 0 and 48 hours. The distance of cell migration was calculated by subtracting the wound width at each time point from the wound width at 0 hour. Three independent assays were performed.
Western blot
Cells were lysed in RIPA lysis buffer (Beyotime, Haimen, China) and nuclear proteins were extracted using lysis buffer (Beyotime); all procedures were performed according to the manufacturer’s protocols. Subsequently, lysates were boiled in 5× SDS-PAGE loading buffer for 10 minutes, resolved by 8% SDS-PAGE, and transferred to nitrocellulose membranes. After blocking, membranes were probed with appropriate antibodies and detected with the ECL™ Western blotting system (Merck, Darmstadt, Germany). Densitometric quantifications were performed using ImageJ image analysis software.
RNA fluorescence in situ hybridization (FISH)
Oligonucleotide primers (F:5′-CTACAAGCAAGATGGCGGAAGAGG-3′, R: 5′-GTAGTCTCCTCGTCCTCCTGGTTC-3′) were used for lncRNA FBXL19-AS1 FISH probe amplification. Briefly, cells were fixed in 4% formaldehyde for 30 minutes and then washed with PBST. The fixed cells were treated with pepsin and dehydrated through ethanol. The cells were incubated for 1 hour with 100 nM of the FISH probe in hybridization buffer. After hybridization, the slide was washed, dehydrated, and mounted with ProLong® Gold Antifade Reagent with DAPI (Cell Signaling Technology, Danvers, MA, USA; #8961) with DAPI for detection. The slides were visualized for immunofluorescence with an Olympus microscope; FV1200 (Olympus, Tokyo, Japan).
Dual luciferase reporter assay
MG63 and 143B cells were seeded at 3×105 cells/well in 6-well plates and allowed to settle overnight. The next day, the cells were co-transfected with pmirGLO-FBXL19-AS1-WT or -MUT reporter plasmids and miR-346 mimic. Relative luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized against Renilla luciferase activity 24 hours after transfection.
Statistical analysis
Statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Differences between 2 groups were assessed using two-tailed Student’s t-tests. Each experiment was performed at least 3 times. Results are displayed as mean ± SD. A P-value <0.05 was considered statistically significant.
Ethics approval and informed consent
Clinical protocols were approved by local regulatory authorities and the study was approved by the ethics committee of Guizhou Provincial People’s Hospital. All patients signed written informed consent before randomization, giving permission to the study team to use biological material for research purposes. The clinical investigations have been conducted relying on the Department of Orthopedics, Guizhou Provincial People’s Hospital’s direction. All animal experiments were approved by the ethics committee of Guizhou Medical University and were performed according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and followed the guidelines of the China Animal Welfare Act.
Results
lncRNA FBXL19-AS1 was upregulated in OS tissues and cell lines
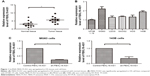
To test whether FBXL19-AS1 plays an important role in OS carcinogenesis, we first measured the level of FBXL19-AS1 expression in 5 human OS cell lines and a normal osteoblast cell line hFOB1.19 by qRT-PCR. The results showed that FBXL19-AS1 was significantly upregulated in OS cells compared with hFOB1.19 cells (Figure 1A, P<0.05). Among the investigated cell lines, MG63 and 143B cells had higher FBXL19-AS1 expression and were chosen for further experiments. Then, FBXL19-AS1 expression was investigated in 12-paired OS and paracancerous tissues; these results showed that FBXL19-AS1 was significantly upregulated in OS compared with normal tissues (Figure 1B, P<0.05). Stable FBXL19-AS1 knockdown was established in MG63 and 143B cells using a lentiviral delivery system, and we confirmed FBXL19-AS1 downregulation in these lines (Figure 1C, P<0.05).
lncRNA FBXL19-AS1 promoted OS cell proliferation in vitro
FBXL19-AS1 knockdown suppressed OS cell proliferation, as measured by the CCK-8 assay. The result showed lower proliferation ability in both cell lines following FBXL19-AS1 knockdown (Figure 2A, P<0.05). We then performed colony formation assays using MG63 and 143B cells to explore the effect of FBXL19-AS1 on colony formation. These assays showed lower colony formation rates in both cell lines following FBXL19-AS1 knockdown (Figure 2B–C, P<0.05). Flow cytometry was used to explore whether FBXL19-AS1 promoted OS cell proliferation by regulating the cell cycle. FBXL19-AS1 knockdown induced a dramatic alteration in the cell cycle distribution of both MG63 and 143B cells: the fraction of cells in G1 phase increased, while the fraction of cells in S phase decreased compared with controls (Figure 2D–E, P<0.05). Western blot analysis showed that FBXL19-AS1 knockdown downregulated cyclin D1, CDK2, and CDK4, which promote cell cycle progression (Figure 2F–G).
lncRNA FBXL19-AS1 promoted OS tumor growth in vivo
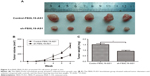
To further explore whether FBXL19-AS1 played a role in OS proliferation in vivo, subcutaneous xenograft models of human OS cells in BALB/c nude mice were established. MG63 cells transfected with sh-FBXL19-AS1 or sh-NC were subcutaneously injected into BALB/c nude mice. The FBXL19-AS1 knockdown group showed a slower increase in tumor diameter and volume and less weight loss in the mice compared with controls (Figure 3A–C, P<0.05).
lncRNA FBXL19-AS1 promoted OS cell migration and invasion in vitro
We next performed wound healing and transwell invasion assays using MG63 and 143B cells to explore the effect of FBXL19-AS1 on cell migration and invasion. A scratch wound healing experiment determined the extent of cell migration by measuring wound narrowing and closure at one time points of 48 hours. The wound area was larger in the sh-FBXL19-AS1 group compared to the sh-NC group (Figure 4A–D, P<0.05). Inconsistently, transwell migration assays showed that FBXL19-AS1 knockdown greatly inhibited the migration of MG63 and 143B cells (Figure 4E–F, P<0.05). We further employed the matrigel invasion assay and found that FBXL19-AS1 knockdown led to a >2-fold reduction in the invasive properties of MG63 and 143B cells (Figure 4G–H, P<0.05). These assays suggested that FBXL19-AS1 knockdown significantly lowered the migratory and invasive ability of both cell lines.
miR-346 was a FBXL19-AS1 target
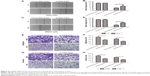
Many lncRNAs have been found to function as ceRNAs that regulate target genes by competitively binding miRNAs. Thus, we next determined whether FBXL19-AS1 operates as a ceRNA. Using starBASE 2.0, we found that miR-346 could potentially bind FBXL19-AS1 (Figure 5A, P<0.05). FISH staining showed the localization of FBXL19-AS1 in MG63 and 143B cells (Figure 5B, P<0.05). RT-PCR results indicated that FBXL19-AS1 was primarily localized within the cytosol (Figure 5C–D, P<0.05). Furthermore, FBXL19-AS1 knockdown upregulated miR-346 expression in MG63 and 143B cells (Figure 5E, P<0.05). The miR-346 expression levels in OS cell lines and clinical sample tissues were determined by the qRT-PCR assay. The results show that the expression level of miR-346 in the OS cell lines and tissues was lower than the normal esophageal epithelial cell line and the adjacent tissues (Figure 5F–G, P<0.05). Pearson’s correlation analysis revealed a negative association between miR-346 and FBXL19-AS1 expression in OS cell lines and OS tissues (Figure 5H–I, P<0.05). We next constructed luciferase reporters containing the miRNA binding sites from FBXL19-AS1. After co-transfecting cells with pmirGLO-FBXL19-AS1-WT or -MUT reporter plasmids, we found that miR-346 overexpression reduced the relative luciferase activity from pmirGLO-FBXL19-AS1-WT, while there was no significant reduction in the luciferase activity from pmirGLO-FBXL19-AS1-MUT (Figure 5J, P<0.05).
miR-346 mediates the pro-proliferation roles of FBXL19-AS1 in OS cells
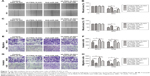
The previous study showed that FBXL19-AS1 was upregulated in OS tissues and cell lines, and our data showed that miR-346 was downregulated. Additionally, FBXL19-AS1 knockdown inhibited the growth of OS cells. Subsequently, we verified the pro- and anti-tumor effects of FBXL19-AS1 and miR-346, respectively, on OS cell lines by co-transfecting anti-miR-346, sh-NC, and sh-FBXL19-AS1. Co-transfecting cells with anti-miR-346 rescued the inhibitory effect of sh-FBXL19-AS1 on cell proliferation (Figure 6A, P<0.05) and colony formation (Figure 6B and C, P<0.05;). These results indicated that reducing miR-346 expression could recover the tumor-suppressive effects of FBXL19-AS1 knockdown in OS cell lines.
miR-346 mediates the pro-migration and -invasion roles of FBXL19-AS1 in OS cells
Next, we investigated whether FBXL19-AS1 promoted migration and invasion in OS cells through miR-346. Functionally, wound healing assays showed a lower migratory ability in both cell lines following co-transfection of anti-miR-346 and sh-FBXL19-AS1 (P<0.05; Figure 7A–D). Furthermore, transwell assays indicated that inhibiting miR-346 promoted the invasion and migration of sh-FBXL19-AS MG63 and 143B cells (P<0.05; Figure 7E–H).
Discussion
Recent studies have revealed that lncRNAs participate in the initiation and progression of various cancers, including breast,7 gastric,8 pancreatic,9 and OS.10 In fact, several lncRNAs have been reported to be involved in OS progression. For example, Zhao et al found that a novel antisense lncRNA HNF1A-AS1 was overexpressed in OS and increased cell proliferation through the Wnt/β-catenin signaling pathway.11 Overexpression of lncRNA MFI2 promotes cell growth and suppresses apoptosis in human OS. Sun et al found that lncRNA FGFR3-AS1 promotes OS progression through FGFR3 activation.12 In the previous study, we detected that a novel lncRNA, FBXL19-AS1, was overexpressed in OS tissues and cell lines. The function of FBXL19-AS1 in OS was subsequently investigated in this study. Our data indicated that FBXL19-AS1 knockdown inhibited OS cell proliferation, migration, and invasion in vitro, as well as tumor growth in vivo.
As to the post-transcriptional regulatory mechanism of FBXL19-AS1, lncRNAs have miRNA responsive elements that act as miRNA sponges to control the endogenous miRNAs that are available to bind target mRNAs, thus reducing the repression of these mRNAs.5 Many lncRNAs have been shown to play roles in tumorigenesis and malignant progression by interfering with miRNA pathways as ceRNAs. lncRNAs have been shown to widely regulate gene expression through different mechanisms, such as pre-transcription, transcription and post-transcription, which mainly depend on their cellular location. Nuclear lncRNAs always regulate pre-transcription or transcription, while cytoplasmic lncRNAs often function as a ceRNAs that sponge miRNAs, thus indirectly regulating target mRNA expression at the post-transcriptional level. In this study, we found that FBXL19-AS1 was mainly cytosolic through RNA FISH and cell fractionation assays, which were qualitative and quantitative tests, respectively, and suggested FBXL19-AS1 regulated mRNAs at the post-transcriptional level. Thus, we suspect it works through the ceRNA mechanism. For example, Xiao et al found that lncRNA-UCA1 regulated miR-16 in chronic myeloid leukemia cells by acting as a ceRNA.13 Liu et al reported that lncRNA SPRY4-IT1 sponges miR-101-3p to promote bladder cancer cell proliferation and metastasis by upregulating EZH2.14 Gao et al reported that lncRNA PVT1 promotes cervical cancer progression by regulating of miR-424.15 Finally, Cai et al reported that lncRNA SNHG16 promotes breast cancer development by acting as a molecular sponge to regulate miR-98/E2F5.16
In this study, we first found that lncRNA FBXL19-AS1 could promote OS cell proliferation, migration, and invasion. An analysis of bioinformatics databases was then conducted to search for miRNAs that have potential binding sites for FBXL19-AS1 in their 3′UTR. This analysis suggested a potential binding partner in miR-346, which was subsequently demonstrated to directly target FBXL19-AS1 in MG63 and 143B cells. However, there were few reports regarding miR-346 in OS. This study showed that miR-346 expression was significantly decreased in OS tissues and cell lines, and negatively correlated with FBXL19-AS1 expression. Furthermore, inhibiting miR-346 rescued the effect of sh-FBXL19-AS1 on OS cell proliferation, migration, and invasion, which suggested an antagonistic role between FBXL19-AS1 and miR-346 in OS. Additionally, direct binding between FBXL19-AS1 and the 3′UTR of miR-346 was validated by the dual luciferase reporter assay. Taken together, these data revealed that lncRNA FBXL19-AS1 could effectively sponge miR-346 to promote OS progression.
Conclusion
We identified the highly expressed oncogenic lncRNA FBXL19-AS1, which plays a crucial role in OS progression. Furthermore, our study sheds light on the role of lncRNA FBXL19-AS1/miR-346 in OS, for the first time, and reveals that FBXL19-AS1 could sponge miR-FBXL19-AS1 to promote OS progression. Thus, this molecule is a novel therapeutic target for OS.
Acknowledgments
This study was supported by research funding from National Natural Science Foundation of China (81560269), Guizhou Provincial Department of Science and Technology, J (2015) 2003, G (2014) 7020, Guizhou Provincial People’s Club Hall, (2013) 05, and the Doctor’s Funding from Guizhou Provincial People’s Hospital GZSYBS (2015) 10. We thank James P Mahaffey, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Worku T, Bhattarai D, Ayers D, et al. Long Non-Coding RNAs: the New Horizon of Gene Regulation in Ovarian Cancer. Cell Physiol Biochem. 2017;44(3):948–966. | ||
Wang H, Yu Y, Fan S, Luo L. Knockdown of Long Noncoding RNA TUG1 Inhibits the Proliferation and Cellular Invasion of Osteosarcoma Cells by Sponging miR-153. Oncol Res. 2018;26(5):665–673. | ||
Yu X, Zheng H, Chan MT, Wu WKK. BANCR: a cancer-related long non-coding RNA. Am J Cancer Res. 2017;7(9):1779–1787. | ||
Wang G, Cui T, Sun L, Peng N, Yang C. Long noncoding RNA LeXis promotes osteosarcoma growth through upregulation of CTNNB1 expression. Am J Cancer Res. 2017;7(7):1577–1587. | ||
Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8(10):17110–17116. | ||
Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. | ||
Peng F, Wang JH, Fan WJ, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2017;27:1062–1074. | ||
Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. | ||
Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233(5):4044–4055. | ||
Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382(2):137–146. | ||
Zhao H, Hou W, Tao J, et al. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Transl Res. 2016;8(8):3503–3512. | ||
Sun J, Wang X, Fu C, et al. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43(5):427–436. | ||
Xiao Y, Jiao C, Lin Y, et al. LncRNA UCA1 Contributes to Imatinib Resistance by Acting as a ceRNA Against miR-16 in Chronic Myeloid Leukemia Cells. DNA Cell Biol. 2017;36(1):18–25. | ||
Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. | ||
Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long Noncoding RNA PVT1 Facilitates Cervical Cancer Progression via Negative Regulating of miR-424. Oncol Res. 2017;25(8):1391–1398. | ||
Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.