Back to Journals » OncoTargets and Therapy » Volume 14
LncRNA DDX11-AS1 Exerts Oncogenic Roles in Glioma Through Regulating miR-499b-5p/RWDD4 Axis
Authors Zheng Y, Xie J, Xu X, Yang X, Zhou Y, Yao Q, Xiong Y
Received 26 August 2020
Accepted for publication 9 December 2020
Published 8 January 2021 Volume 2021:14 Pages 157—164
DOI https://doi.org/10.2147/OTT.S278986
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Yanyan Zheng,1 Jing Xie,2 Xiaomin Xu,1 Xiaoguo Yang,1 Yi Zhou,3 Qiong Yao,1 Ye Xiong4
1Department of Neurology, Wenzhou Third Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, Wenzhou 325000, People’s Republic of China; 2Department of Dermatology, Wenzhou Third Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, Wenzhou 325000, People’s Republic of China; 3Department of Neurosurgery, Wenzhou Third Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, Wenzhou 325000, People’s Republic of China; 4Department of Neurosurgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Correspondence: Ye Xiong
Department of Neurosurgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Email [email protected]
Background: Long noncoding RNAs (lncRNA) exert essential functions during tumorigenesis. However, how lncRNAs participate in glioma development remains poorly researched. This study aimed to determine how DDX11-AS1 affects glioma progression.
Methods: Gene expression was analyzed by qRT-PCR. Survival rate curve was plotted in 56 glioma patients. Loss-of-function assays were performed to analyze proliferation, migration, and invasion through CCK8, colony formation, and transwell assays. Luciferase assay and RNA pulldown assays were conducted to illustrate the underlying molecular mechanism.
Results: DDX11-AS1 expression was upregulated in glioma tissues and cells. DDX11-AS1 overexpression was linked with poor prognostic value. DDX11-AS1 knockdown suppressed proliferation, migration, and invasion while inducing apoptosis. DDX11-AS1 interacted with miR-499b-5p to eliminate it, leading to upregulation of RWDD4 expression. RWDD4 was upregulated in glioma while miR-499b-5p was downregulated.
Conclusion: DDX11-AS1 upregulation promotes glioma progression through acting as a competing endogenous RNA for miR-499b-5p to upregulate RWDD4.
Keywords: DDX11-AS1, miR-499b-5p, RWDD4, glioma
Introduction
Glioma derives from the central nervous system and is a rather aggressive cancer with a high mortality.1 Glioma could be classified into four grades, including I, II, III, and IV, according to cellular behaviors and histopathological characteristics.2 In particular, glioblastoma is the most malignant subtype of glioma.3 Radiotherapy and chemotherapy combined with surgery are the current main therapeutic methods against glioma.4 Nevertheless, the overall survival rate of glioma patients is very low.5 Therefore, it is important to understand the molecular mechanism underlying glioma progression.
Long noncoding RNAs (lncRNA) represent a kind of RNAs with over 200 nucleotides in length and without protein-coding potential.6 Recently, a growing number of studies indicate that lncRNAs play critical roles in several biological processes, such as immune and cancer.7,8 Aberrant expression of lncRNAs may lead to dysregulation of proliferation, survival, and metastasis of tumor cells.9 For example, lncRNA JPX is upregulated in lung cancer and promotes tumor development through targeting miR-145/CCND2 axis.10 LncRNA OIP5-AS1 overexpression contributes to proliferation, migration, and invasion in pancreatic ductal adenocarcinoma.11 In addition, LINC01614 upregulation sponges miR-383 to promote glioma growth and invasion by affecting ADAM12 expression.12 Therefore, illustrating the biological roles and molecular mechanisms of lncRNAs is of important significance.
DDX11-AS1 has been researched and identified as an oncogenic lncRNA in several cancers, such as gastric cancer, bladder cancer, and esophageal cancer.13–15 Its role in glioma remains undefined. In the current study, DDX11-AS1 was found to be upregulated in glioma and correlated with poor prognosis. DDX11-AS1 knockdown impaired proliferation, migration, and invasion in glioma cells. DDX11-AS1 was identified to sponge miR-499b-5p and upregulate RWDD4. In conclusion, our findings suggest that DDX11-AS1/miR-499b-5p/RWDD4 axis plays essential roles in regulating glioma progression.
Patients and Methods
Patients’ Tissues
Fifty-six glioma tissues and adjacent normal tissues were collected from the Frist Affiliated Hospital of Wenzhou Medical University. All samples were diagnosed by at least two pathologists. Patients were not treated by chemotherapy or radiotherapy prior to surgery. Tissues were stored in nitrogen until use. All patients provided written informed consents. This study was approved by the Ethics Committee of the Frist Affiliated Hospital of Wenzhou Medical University. All experiments were performed in accordance with the Declaration of Helsinki.
Cell and Transfection
Glioma cell lines and normal human astrocytes (NHAs) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured using RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 100 U/mL penicillin/streptomycin (Invitrogen). siRNAs, miR-499b-5p mimics, miR-499b-5p inhibitors and corresponding negative controls were purchased from GenePharma (Shanghai, China). Transfection was carried out through Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
qRT-PCR
RNA was isolated from tissues and cells using Trizol reagents and utilized for reverse transcription using the cDNA synthesis kits (Takara, Dalian, Liaoning, China). qPCR was conducted using the TB Green™Premix Ex Taq™ II (TaKaRa). To analyze miRNA expression, cDNA was generated using the miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN) and qPCR was conducted through miRcute Plus miRNA qPCR Kit (SYBR Green). ACTB or U6 was used for normalized control. Relative expression was calculated according to the 2−ΔΔCt methods.
Cell Proliferation
As for CCK8 (Dojindo) assay, cells were plated into 96-well plates and cultured for indicated days. Then, CCK8 solution was added and incubated for 2 h. Finally, the absorbance at 450 nm was measured using a microplate reader. For colony formation assay, cells were seeded into 6-well plates and cultured for 14 days. Then, clones were fixed with methanol and stained with 0.1% crystal violet. Colony numbers were counted.
Transwell Assays
Cells were seeded into the upper chamber (pre-coated with Matrigel for detection of invasion) in 200 μL serum-free medium. The lower chamber was filled with 600 μL serum-containing medium. After cultured for 48 h, the migrated or invaded cells in the lower member were fixed with methanol and stained with crystal violet (0.1%). Then, cell images were acquired using a microscope.
Caspase 3 Activity Test
The apoptosis was analyzed by using a Caspase3 activity assay kit (Abcam, Cambridge, UK) according to the manufacturer’s instructions.
Dual-Luciferase Activity Assay
The predicted regions of DDX11-AS1 and RWDD4 containing miR-499b-5p-binding elements were inserted into pGL3 reporter plasmid. Then, luciferase reporter plasmid and miR-499b-5p mimics were co-transfected into glioma cells. After 48 h, the luciferase activity was measured using Promega Luciferase Reporter detection kits (Promega).
Pulldown Assay
Biotin-labeled DDX11-AS1 (full length) and negative control (antisense of DDX11-AS1) were obtained and incubated with cell lysates. Then, magnetic beads (Invitrogen, Pudong, Shanghai, China) were added and incubated for 4 h. The beads were washed and eluted RNAs were isolated and analyzed by qRT-PCR.
Statistical Analysis
Graphpad Prism 6 software was used for statistical analysis. Kaplan–Meier methods with Log-rank tests were used for overall survival analysis. Student’s t-test and a one-way ANOVA were conducted to determine significant differences. P values <0.05 were considered as statistically significant.
Results
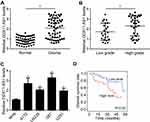
DDX11-AS1 Was Upregulated in Glioma
To explore the potential function of DDX11-AS1, its expression in glioma was analyzed. qRT-PCR result showed that DDX11-AS1 level was raised in glioma tissues (Figure 1A). Moreover, DDX11-AS1 levels were positively correlated with tissue grade (Figure 1B). Similarly, it was observed that DDX11-AS1 expression was higher in glioma cell lines than that in NHAs (Figure 1C). Next, glioma tissues were divided into two groups based on DDX11-AS1 median value. We observed that DDX11-AS1 high expression was correlated with low survival rate (Figure 1D).
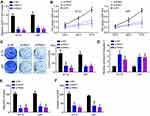
DDX11-AS1 Knockdown Inhibited Glioma Progression
Afterwards, A172 and U87 cells were selected to investigate the function of DDX11-AS1. Two independent siRNAs were used to knock DDX11-AS1 down in A172 and U87 cells (Figure 2A). CCK8 assay showed that DDX11-AS1 knockdown significantly inhibited proliferation rate in A172 and U87 cells (Figure 2B). Consistently, knockdown of DDX11-AS1 depleted glioma cells formed less clones (Figure 2C). Next, the activity of Caspase 3 was measured. Results indicated that DDX11-AS1 knockdown upregulated the activity of Caspase 3 (Figure 2D), suggesting DDX11-AS1 knockdown induces apoptosis. Besides, transwell assay was conducted. It was found that DDX11-AS1 knockdown led to decreased numbers of migrated and invaded cells (Figure 2E and F).
DDX11-AS1 Exerted as the ceRNA for miR-499b-5p
qRT-PCR result indicated that DDX11-AS1 was mainly expressed in the cytoplasm of A172 cells (Figure 3A). We performed bioinformatics analysis using miRBD and identified several miRNA candidates. However, only miR-499b-5p ranked top and has been reported to be sponged by DDX11-AS1 in bladder cancer.14 Thus, we focused on miR-499b-5p and constructed wide-type (WT) and mutant (MUT) luciferase reporters (Figure 3B). Luciferase reporter assay suggested that miR-499b-5p mimics suppressed the activity of WT-DDX11-AS1 reporter (Figure 3C). RNA pulldown assay also confirmed the interaction between DDX11-AS1 and miR-499b-5p (Figure 3D). Additionally, it was observed that DDX11-AS1 was negatively correlated with miR-499b-5p level in glioma tissues (Figure 3E). Next, we found that DDX11-AS1 knockdown caused upregulation of miR-499b-5p expression in A172 and U87 cells (Figure 3F). Besides, miR-499b-5p was downregulated in glioma tissues (Figure 3G).
RWDD4 Was the Target of miR-499b-5p
Next, the targets of miR-499b-5p were analyzed by bioinformatics analysis through TargetScan and miRDB. RWDD4 ranked top and was selected for further validation. We constructed luciferase reporter vectors (Figure 4A). Similarly, luciferase reporter assay showed that miR-499b-5p mimics inhibited the activity of WT-RWDD4 vector (Figure 4B). We observed that RWDD4 was upregulated in glioma tissues (Figure 4C and D). And RWDD4 level was reversely correlated with miR-499b-5p in glioma tissues (Figure 4E). Interestingly, we noticed that miR-499b-5p mimics or DDX11-AS1 siRNA transfection suppressed the expression of RWDD4 (Figure 4F). Moreover, miR-499b-5p inhibitors reversed the effect of DDX11-AS1 siRNA (Figure 4F), indicating that DDX11-AS1was the sponge for miR-499b-5p to upregulate RWDD4. Finally, it was observed that DDX11-AS1 was positively correlated with RWDD4 expression in glioma tissues (Figure 4G).
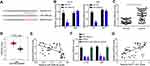 |
Figure 4 RWDD4 was the target of miR-499b-5p. (A) The binding site between miR-499b-5p and RWDD4. (B) Luciferase reporter assay. (C) RWDD4 expression was upregulated in glioma tissues through qRT-PCR assay. (D) TCGA data in GEPIA database (http://gepia.cancer-pku.cn/detail.php?gene=RWDD4) indicated that RWDD4 was upregulated in glioma tissues. (E) Expression correlation between miR-499b-5p and RWDD4 in glioma tissues. (F) Relative expression of RWDD4 after transfection with indicated plasmids. (G) Expression correlation between DDX11-AS1 and RWDD4 in glioma tissues. *P<0.05. |
DDX11-AS1 Promoted Glioma Development Through Regulating miR-499b-5p/RWDD4 Axis
To further validate the physiological roles of DDX11-AS1/miR-499b-5p/RWDD4 in glioma, rescue assays were designed by transfection with miR-499b-5p inhibitors or pcDNA3-RWDD4. CCK8 assay showed that miR-499b-5p inhibition or RWDD4 restoration rescued the proliferation ability of glioma cells (Figure 5A). Similarly, the apoptosis, migration, and invasion were also reversed by miR-499b-5p inhibition or RWDD4 restoration (Figure 5B–D). Therefore, DDX11-AS1 promotes glioma progression through targeting miR-499b-5p/RWDD4 axis.
Discussion
Glioma is a very malignant brain tumor and characterized with a poor prognosis. Its underlying molecular mechanism remains unclear. In this study, we investigated how lncRNA regulates glioma progression. We showed that DDX11-AS1 level was raised in glioma and positively correlated with clinical grade. Besides, DDX11-AS1 silencing suppressed glioma proliferation, migration, and invasion while causing cell death. Mechanistically, DDX11-AS1 was demonstrated to sponge miR-499b-5p to facilitate RWDD4 expression. Our work expounds the important roles of DDX11-AS1/miR-499b-5p/RWDD4 axis in promoting glioma development.
There is a close link between lncRNA and cancer. The importance of lncRNA in glioma has also been validated. For example, LINC00174 enhances the chemoresistance of glioma cells and promotes glioma progression through miR-138-5p/SOX9 pathway.16 LncRNA HOXA-AS2 promotes glioma growth and migration through affecting RND3 level.17 Besides, lncRNA FLVCR1-AS1 contributes to glioma proliferation, migration, and invasion by regulating miR-4731-5p/E2F2 signaling.18 Previous researches indicate that DDX11-AS1 is a potential oncogene in several cancers. DDX11-AS1 promotes the malignant behaviors of gastric cancer, bladder cancer, lung cancer, osteosarcoma, esophageal cancer, colorectal cancer, and liver cancer.13–15,19,20 However, whether DDX11-AS1 regulates glioma is not investigated. In our work, we demonstrated that DDX11-AS1 expression was upregulated in glioma tissues. DDX11-AS1 upregulation predicted poor prognosis. Moreover, we illustrated that DDX11-AS1 promoted proliferation, migration, invasion, and survival of glioma, suggesting it is a new oncogene in glioma.
LncRNAs have been found to utilize several mechanisms to exert functions, such as regulating translation or transcription and epigenetic modulation.21 LncRNA works as a miRNA sponge to regulate tumorigenesis.21 For example, LncRNA LINC00689 sponges miR-526b to promote gastric cancer development.22 LncRNA MT1JP sponges miR-214 to inhibit breast cancer progression.23 DDX11-AS1 has also been reported to sponge several miRNAs, such as miR-499b-5p, miR-873, and miR-326.13,14,19 In our study, we found that DDX11-AS1 was distributed in the cytoplasm of glioma cells. We also demonstrated that DDX11-AS1 was the sponge for miR-499b-5p by luciferase reporter assay and RNA pulldown assay. Only one study showed that miR-499b-5p suppresses bladder cancer development.14 Its function in other cancer is fully unknown. Our data showed that miR-499b-5p was downregulated in glioma tissues and its expression was restricted by DDX11-AS1. Besides, we showed that miR-499b-5p inhibitors promoted glioma progression. Thus, our study demonstrated miR-499b-5p as a tumor suppressor in glioma.
The lncRNA-miRNA-mRNA regulatory axis is widely acknowledged in cancer.8,18 Thus, we performed bioinformatics analysis to search for the target of miR-499b-5p. We identified RWDD4 as the most potential target. Through luciferase reporter assay, the interaction between miR-499b-5p and RWDD4 was demonstrated. Our data indicated that RWDD4 was inhibited by miR-499b-5p and upregulated by DDX11-AS1. Moreover, DDX11-AS1 promoted RWDD4 expression by restricting miR-499b-5p. A previous work indicates RWDD4 as an oncogene in bladder cancer.24 To date, no other study reports its other function. And whether RWDD4 plays a similar role in glioma needs to be investigated. Our data showed that RWDD4 expression was upregulated in glioma tissues. Moreover, RWDD4 overexpression rescued the proliferation, migration, and invasion abilities of glioma cells, suggesting RWDD4 is a critical oncogene in glioma.
In summary, we elucidated that DDX11-AS1 promotes glioma progression through acting as a sponge for miR-499b-5p to facilitate RWDD4 expression. Our findings provide a potential therapeutic target for glioma treatment. However, there are some limitations in our study. For instance, it will be more representative to use patient-derived glioma cells for functional experiments. Besides, in vivo experiments will be important to confirm the roles of DDX11-AS1.
Acknowledgments
The project was supported by Zhejiang Provincial Natural Science Foundation (LGF18H090007), Basic Research Programs of Science and Technology Department of Wenzhou (Y2020219) and Youth Talents Plan of Science and Technology Project of Zhejiang Provincial Health Commission (2019RC276).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi:10.1056/NEJMra0708126
2. Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. doi:10.1016/j.semcancer.2017.11.010
3. Perry A, Wesseling P. Histologic classification of gliomas. Handb Clin Neurol. 2016;134:71–95.
4. Ferris SP, Hofmann JW, Solomon DA, Perry A. Characterization of gliomas: from morphology to molecules. Virchows Arch. 2017;471(2):257–269. doi:10.1007/s00428-017-2181-4
5. Rizzo D, Ruggiero A, Martini M, Rizzo V, Maurizi P, Riccardi R. Molecular biology in pediatric high-grade glioma: impact on prognosis and treatment. Biomed Res Int. 2015;2015:215135. doi:10.1155/2015/215135
6. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46.
7. Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi:10.1038/ni.3712
8. Han B, Ge Y, Cui J, Liu B. Down-regulation of lncRNA DNAJC3-AS1 inhibits colon cancer via regulating miR-214-3p/LIVIN axis. Bioengineered. 2020;11(1):524–535. doi:10.1080/21655979.2020.1757224
9. Lian Y, Yan C, Lian Y, et al. Long intergenic non-protein-coding RNA 01446 facilitates the proliferation and metastasis of gastric cancer cells through interacting with the histone lysine-specific demethylase LSD1. Cell Death Dis. 2020;11(7):522. doi:10.1038/s41419-020-2729-0
10. Jin M, Ren J, Luo M, et al. Long non-coding RNA JPX correlates with poor prognosis and tumor progression in non-small-cell lung cancer by interacting with miR-145-5p and CCND2. Carcinogenesis. 2020;41(5):634–645. doi:10.1093/carcin/bgz125
11. Wu L, Liu Y, Guo C, Shao Y. LncRNA OIP5-AS1 promotes the malignancy of pancreatic ductal adenocarcinoma via regulating miR-429/FOXD1/ERK pathway. Cancer Cell Int. 2020;20:296. doi:10.1186/s12935-020-01366-w
12. Wang H, Wu J, Guo W. SP1-mediated upregulation of lncRNA LINC01614 functions a ceRNA for miR-383 to facilitate glioma progression through regulation of ADAM12. Onco Targets Ther. 2020;13:4305–4318. doi:10.2147/OTT.S242854
13. Song W, Qian Y, Zhang MH, et al. The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24(6):3049–3061. doi:10.26355/eurrev_202003_20669
14. Li Q, Wang S, Wu Z, Liu Y. DDX11-AS1exacerbates bladder cancer progression by enhancing CDK6 expression via suppressing miR-499b-5p. Biomed Pharmacother. 2020;127:110164. doi:10.1016/j.biopha.2020.110164
15. Zhang S, Jiang H, Xu Z, et al. The resistance of esophageal cancer cells to paclitaxel can be reduced by the knockdown of long noncoding RNA DDX11-AS1 through TAF1/TOP2A inhibition. Am J Cancer Res. 2019;9(10):2233–2248.
16. Li B, Zhao H, Song J, Wang F, Chen M. LINC00174 down-regulation decreases chemoresistance to temozolomide in human glioma cells by regulating miR-138-5p/SOX9 axis. Hum Cell. 2020;33(1):159–174. doi:10.1007/s13577-019-00281-1
17. Wu L, Zhu X, Song Z, et al. Long non-coding RNA HOXA-AS2 enhances the malignant biological behaviors in glioma by epigenetically regulating RND3 expression. Onco Targets Ther. 2019;12:9407–9419. doi:10.2147/OTT.S225678
18. Yan Z, Zhang W, Xiong Y, Wang Y, Li Z. Long noncoding RNA FLVCR1-AS1 aggravates biological behaviors of glioma cells via targeting miR-4731-5p/E2F2 axis. Biochem Biophys Res Commun. 2020;521(3):716–720. doi:10.1016/j.bbrc.2019.10.106
19. Tian JB, Cao L, Dong GL. Long noncoding RNA DDX11-AS1 induced by YY1 accelerates colorectal cancer progression through targeting miR-873/CLDN7 axis. Eur Rev Med Pharmacol Sci. 2019;23(13):5714–5729. doi:10.26355/eurrev_201907_18309
20. Li Y, Zhuang W, Huang M, Li X. Long noncoding RNA DDX11-AS1 epigenetically represses LATS2 by interacting with EZH2 and DNMT1 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;514(4):1051–1057. doi:10.1016/j.bbrc.2019.05.042
21. Meng X, Deng Y, Lv Z, et al. LncRNA SNHG5 promotes proliferation of glioma by regulating miR-205-5p/ZEB2 axis. Onco Targets Ther. 2019;12:11487–11496. doi:10.2147/OTT.S228439
22. Yin G, Tian P, BuHe A, Yan W, Li T, Sun Z. LncRNA LINC00689 promotes the progression of gastric cancer through upregulation of ADAM9 by sponging miR-526b-3p. Cancer Manag Res. 2020;12:4227–4239. doi:10.2147/CMAR.S231042
23. Ouyang Q, Cui Y, Yang S, et al. lncRNA MT1JP suppresses biological activities of breast cancer cells in vitro and in vivo by regulating the miRNA-214/RUNX3 Axis. Onco Targets Ther. 2020;13:5033–5046. doi:10.2147/OTT.S241503
24. Hou Y. MiR-506 inhibits cell proliferation, invasion, migration and epithelial-to-mesenchymal transition through targeting RWDD4 in human bladder cancer. Oncol Lett. 2019;17(1):73–78. doi:10.3892/ol.2018.9594
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.