Back to Journals » OncoTargets and Therapy » Volume 12
LncRNA CASC15 Functions As An Unfavorable Predictor Of Ovarian Cancer Prognosis And Inhibits Tumor Progression Through Regulation Of miR-221/ARID1A Axis
Authors Shi Y, Gao S, Zheng Y, Yao M, Ruan F
Received 18 June 2019
Accepted for publication 27 August 2019
Published 23 October 2019 Volume 2019:12 Pages 8725—8736
DOI https://doi.org/10.2147/OTT.S219900
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Yin Shi, Shanshan Gao, Ying Zheng, Mukun Yao, Fan Ruan
Department of Gynecology, Tongde Hospital of Zhejiang Province, Hangzhou, Zhejiang Province, People’s Republic of China
Correspondence: Fan Ruan
Department of Gynecology, Tongde Hospital of Zhejiang Province, No. 234 Gucui Road, Xihu District, Hangzhou City, Zhejiang Province, People’s Republic of China
Tel +86 571-89972000
Email [email protected]
Background: LncRNA cancer susceptibility candidate 15 (CASC15) has been demonstrated to act as an oncogene in different cancers; however, its role in ovarian cancer remains elusive.
Methods: Quantitative real-time PCR (qRT-PCR) was performed to examine the expression of lncRNA CASC15. Kaplan–Meier survival analysis was performed to evaluate the prognostic significance of lncRNA CASC15. CCK-8, soft-agar colony-formation, flow cytometry, transwell migration and invasion assays were used to analyze the biological behavior of lncRNA CASC5 in ovarian cancer. Furthermore, the potential mechanism of lncRNA CAC15 was investigated by bioinformatics analysis, luciferase reporter assay, and biotin pull-down assay.
Results: In this study, we found that the expression of CASC15 was lower in ovarian cancer tissues and cells by qRT-PCR. In addition, low expression of CASC15 was closely correlated with advanced TNM stage, moderate/poor differentiation, and larger size. Moreover, Kaplan–Meier survival analysis showed that patients with low CASC15 expression level had poorer overall survival and progression-free survival than those with high CASC15 expression. Meanwhile, ROC analysis found that CASC15 had diagnostic values to distinguish tumor tissues from nontumorous tissues. Overexpression of CASC15 prohibited the malignancy of ovarian cancer cells, including proliferation, colony formation, cell cycle, migration, and invasion, and promoted cell apoptosis. In addition, bioinformatics analysis, luciferase reporter assay, and biotin pull-down assay confirmed that CASC15 straightly interacted with miR-221. We also observed that ARID1A was a downstream target of miR-221 and CASC15 subsequently exerted its tumor-suppressive effects by regulating the expression of ARID1A in ovarian cancer cells.
Conclusion: Overall, this study firstly elucidated that CASC15 could play a tumor-suppressive role in ovarian cancer by the regulation of CASC15/miR-221/ARID1A axis, which may provide a ponderable prognostic biomarker and promising therapeutic target for treatment of patients with ovarian cancer.
Keywords: long non-coding RNA, CASC15, ovarian cancer, miR-221, ARID1A
Introduction
Ovarian cancer is one of the most common gynecological malignancies that has high morbidity and mortality all over the world.1 Although chemotherapy, radiotherapy, target-therapy, and immunological therapy have gained great advance for the therapy of this cancer and new biomarkers for early ovarian cancer diagnose are also developed quickly, the 5-year overall survival (OS) rate of ovarian cancer remains poor due to metastasis, recurrence, and acquired drug resistance.2,3 Thus, there is a compelling clinical demand to understand the molecular mechanisms underlying ovarian cancer pathogenesis, development, and its progression, explore novel biomarkers to predict tumorigenesis, clinical diagnosis, and explore new therapy strategies and targets of ovarian cancer.4
In recent years, long noncoding RNAs (lncRNAs) are non-encoding RNA molecules that are usually longer in length than 200 nucleotides and lack open reading frame (ORF), but they can function as competing endogenous RNAs (ceRNAs) by acting as special miRNAs’ sponges to regulate their targets and be involved in several physiological processes, including the development of many malignant tumors.5–8 LncRNA CASC15, locating on human chromosome 6p22.3, have been demonstrated to act as one novel oncogene to regulate the growth, metastasis and drug resistance in many cancers, such as colorectal cancer, cervical cancer, breast cancer, gastric cancer, tongue squamous carcinoma, hepatocellular carcinoma, and melanoma.9–17 For example, Yu showed that lncRNA cancer susceptibility candidate 15 accelerated the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop.11 Wu also demonstrated that lncRNA CASC15 regulated gastric cancer cell proliferation, migration, and epithelial–mesenchymal transition by targeting Cyclin-dependent kinase inhibitor 1a (CDKN1A) and Zinc finger E-box-binding homeobox 1 (ZEB1).12 However, there are no studies about the function of the lncRNA CASC15 in ovarian cancer. Besides, it has been demonstrated that miR-221 acts as an oncogene to regulate proliferation and apoptosis of ovarian cancer cells.18,19 In addition, it has been reported that ARID1A has been recognized as a well-known tumor suppressor in ovarian cancer.20 However, whether miR-221 could promote progression of ovarian cancer cells via targeting ARID1A remains elusive.
In the present study, for the first time, we found CASC15 was significantly downregulated in ovarian cancer tissues and cells. The low expression of CASC15 was closely associated with shorter OS and progression-free progression (PFS) of patients with ovarian cancer. Moreover, CASC15 was recognized as a ceRNA for miR-221 and could exert its tumor-suppressive effects on cell growth, migration, and invasion through the regulation of ARID1A. Overall, our study firstly reported that CASC15 could act as a tumor suppressor to modulate ovarian cancer development and progression by CASC15/miR-221/ARID1A axis.
Materials And Methods
Clinical Specimens
Paired ovarian cancer tissues and adjacent normal tissues were collected during January 2012–January 2014 from 102 patients who underwent surgery at Tongde Hospital of Zhejiang Province. All obtained specimens were instantly frozen in liquid nitrogen and kept for subsequent use. All patients did not receive any systematic therapy or chemotherapy before surgical resection. The informed consents were signed by all patients for the utilization of tissue samples in research. The Ethics Committee of Tongde Hospital of Zhejiang Province approved this study. Samples were fixed in 10% formalin and embedded in paraffin. H&E staining was performed on sections from embedded samples to differentiate the normal and cancer tissues. The clinicopathological features of all patients are listed in Table 1.
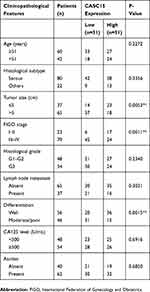 |
Table 1 Association Between CASC15 Expression And Clinicopathological Characteristics Of Patients With Ovarian Cancer |
Cell Culture And Transfection
Six ovarian cancer cell lines (OVCAR3, SKOV3, A2780, HO-8910, Caov3, and CP70) and human ovarian immortalized nontumorigenic ovarian surface epithelial cells (IOSE) were obtained from the American type culture collection (ATCC) (Maryland, USA) and cultured in RPMI 1640 (TransGen, Beijing, China) medium or DMEM (TransGen, Beijing, China) supplemented with 10% fetal bovine serum (Gibco, 10%FBS), 1% penicillin/streptomycin (Sigma, USA). All the cells were maintained in humidified chamber at 37°C with 5% CO2.
miR-221 mimics (catalog number: AM16697), miR-221 inhibitor (anti-miR-221, catalog number: AM16812) as well as the negative control were all obtained from Sigma-Aldrich (Sigma, USA). Cell transfection was performed using Lipofectamine 3000 Reagent (Life Technologies, San Diego, CA, USA) based on the manufacturer’s protocol. The CASC15 sequences were ligated into the pEX-2 vector (pEX-CASC15) and pEX-2 vector-control was used as a negative control (pEX) (Catalog number: pEX-2, Genepharma, China).
RNA Extraction And RT-qPCR
Total RNA of tissues or ovarian cancer cells was extracted by TRIzol reagent (Invitrogen, USA) according to the manufacturer’s guide. RNA quantification was conducted with a NanoDrop ND-3000 spectrophotometer (Life Technologies). cDNA was synthesized and quantitative RT-PCR was performed using ABI 7500 real-time PCR system (Applied Biosystems, USA) to measure the relative expression of target gene according to the manufacture’s protocol. All the primers were purchased by Genecopoeia (Guangzhou, China). Relative expression of target genes and miRNA expression were calculated using 2−ΔΔCt method. The following primers were used in this study: CASC15, forward: 5ʹ-CCCCATGATGGTTCCTCAGTT-3ʹ, reverse 3ʹ-GGAAAGCAAGTGCAGGTTAGTC-5ʹ; miR-221, forward: 5ʹ-AGCTACATTGTCTGCTGGGTTTC-3ʹ; reverse, 5ʹ-GATCCGAAACCCAGCAGACAATGTAGCTTTTTT-3ʹ; GAPDH, forward: 5ʹ-GTCATCCCTGAGCTAGACGG-3ʹ, reverse, 5ʹ-GGGTCTTACTCCTTGGAGGC-3ʹ.
Cell Proliferation Assay
Cell Counting Kit-8 assay (CCK-8, Dojindo; Kumamoto, Japan) was used to detect the cell viability according to the manufacturer’s guide. Briefly, ovarian cancer cells were cultured into 96-well plates at a density of 1000–3000 cells/well. As the indicated time after transfection, the OD value was measured using FLx800 Fluorescence Microplate Reader (Biotek).
Soft-Agar Colony-Formation Assay
CytoSelect™ 96-Well Cell Transformation Assay (Standard Soft Agar kit, Cell Biolabs) was used to detect the colony formation ability according to the manufacturer’s guide. A total of 5000 cells per well of ovarian cancer cells were seeded in 96-well plate and then incubated for 2 weeks. The colonies were then measured with the CyQuant and analyzed using FLx800 Fluorescence Microplate Reader (Biotek).
Detection Of Caspase-3 Activation And Cell Cycle Analysis
Apoptotic activity was detected by active caspase 3 human ELISA kit (Invitrogen Corporation, Camarillo, CA) according to the manufacture’s guide. The detailed experiment procedure was consistent with the previous description.21 The activity was read using a microplate reader at 450 nm using Envision (PerkinElmer). For the cell cycle analysis, cells were stained with propidium iodide (PI) (Sigma) and analyzed by flow cytometry (FACS Calibur, BD Biosciences) as previous description.22
Migration And Invasion Assay
The migration and invasion were measured by transwell chamber without or with Matrigel (8-μm pore size; BD Biosciences, USA) according to the manufacturer’s protocol. The procedure was consistent with the previous description.23 The migrated or invaded cells were stained with 1% crystal violet, photographed using a microscope with a 10× objective (total magnification 100×) and counted. Experiments were performed in triplicate.
Dual-Luciferase Reporter Assay
Bioinformatics tools (Starbasev2.0 and miRcode) were used to analyze the miR-221 binding sites on CASC15 gene as well as the ARID1A binding sites on miR-221. The putative miR-221 target binding sequence in CASC15 and its mutant of the binding sites was amplified by PCR and the PCR products were ligated into pMirReporter plasmid (Sigma, USA) to produce the CASC15-wild-type (wt-CASC15) vector and CASC15-mutated-type (mut-CASC15). Similarly, the ARID1A-wild-type (wt-ARID1A) and ARID1A-mutated-type (mut-ARID1A) reporter vectors were synthesized. Subsequently, mutated or wild-type vector and miR-221 mimics or NC-mimics were cotransfected into HEK-293 cells in 96-well plates for 24 hrs using Lipofectamine 3000 (Thermo Fisher Scientific). The luciferase reporter assay system was used to measure the luciferase activity (Promega, Madison, WI, USA) according to the manufacturer’s protocol as previous description.24
Pull Down Assay With Biotinylated lncRNA CASC15 DNA Probe
Briefly, the DNA fragment with the full-length CASC15 sequence or negative control sequence was PCR amplified by a T7-containing primer and then binded into GV394 (Invitrogen, Shanghai, China). The obtained plasmid DNAs were digested to be linear by restriction enzyme XhoI. Biotin-labeled RNAs were conversely transcribed and then qRT-PCR was used to analyze the target RNA expression according to the method described in the previous study.25
Western Blot Analysis
Cells were harvested and lysed via RIPA lysis buffer added with proteinase inhibitors cocktail (Sigma). Western blot analysis was performed as previous description.26 Briefly, lysed protein was isolated by SDS-PAGE, transferred to PVDF membranes, and incubated with the following primary antibodies: β-actin antibody (1:10,000 dilution, mouse, Sigma); ARID1A antibody (1:500 dilution, Rabbit, Santa Cruz, CA, USA). Following incubation with HRP-conjugated appropriate secondary antibody, the bands were visualized using Pierce™ ECL Western Blotting Substrate (32,109, Thermo Scientific, USA). The signal intensity of protein was further determined by Image J software (Madison, WI, USA).
Statistical Analysis
All data were expressed as the mean ± Standard Deviation (SD). Statistical analysis was performed with SPSS Version 22.0 (SPSS, Chicago, USA) and graphs were plotted with GraphPad Prism 7 (La Jolla, CA, USA). The significance of differences between groups was evaluated by Student’s t-test or one-way analysis of variance (ANOVA). The Kaplan–Meier method test and Log-rank test was performed for analysis of PFS and OS. The relationship between two genes was determined by Pearson correlation analysis. The receiver operating characteristic (ROC) curve was used to analyze the diagnostic value. The difference with P<0.05 was considered to be statistically significant.
Results
LncRNA CASC15 Expression Was Downregulated In Ovarian Cancer And Related With Poor Prognosis Of Ovarian Cancer Patients
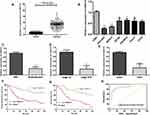
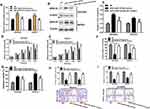
RNA sequencing expression data of CASC15 from the TCGA and the GTEx projects were used to investigate the expression of CASC15 in ovarian cancer by bioinformatic website-“GEPIA” (http://gepia.cancer-pku.cn/). As shown in Figure 1A, the data showed that lncRNA CASC15 was the most downregulated in ovarian cancer tissues among multiple kinds of cancer. As shown in Figure 1B, lncRNA CASC15 was dramatically reduced compared to normal tissues (*P<0.05). To differentiate the ovarian cancer tissue and normal ovarian tissue, we performed H&E staining and representative photographs of H&E staining showed that we could obtain the ovarian cancer tissues and its paired adjacent normal tissues (Figure S1). Then, qRT-PCR assay was applied to analyze the expression of CASC15 in 102 paired ovarian cancer tissues and adjacent normal tissues from our hospital. The results exhibited that CASC15 expression was markedly decreased in ovarian cancer tissues compared with adjacent normal tissues (Figure 2A, P<0.001). In addition, we found that strikingly underexpressed CASC15 could be observed in 6 ovarian cancer cell lines compared to normal human colonic epithelial cell (HcoEpiC) by qRT-PCR (Figure 2B, P<0.05). To better understand clinical significance of CASC15, we showed that CASC15 expression was notably downregulated in ovarian cancer tissues with moderate/poor differentiation (Figure 2C, P<0.01), high clinical stage (Ⅲ–Ⅳ) (Figure 2D, P<0.01), and larger size (>5cm) (Figure 2E, P<0.01) compared to those with well differentiation, low clinical stage, and small size, respectively. To further study the correlation between lncRNA CASC15 expression and clinicopathological characteristics, tumors were classified into low and high CASC15 expression groups according to the median expression of CASC15. As shown in Table 1, low CASC15 expression was significantly associated with tumor size, FIGO stage, and differential by Chi-square test. However, the CASC15 expression levels showed no correlation with other clinical parameters, such as age, histological subtype, histological grade, lymph node metastasis, CA125 level, or ascites. Moreover, Kaplan–Meier analysis displayed that low CASC15 expression strongly shortens PFS (Figure 2F, P<0.01) and OS (Figure 2G, P<0.001) of CRC patients. The diagnostic value of lncRNA CASC15 in the ovarian cancer patients was assessed by ROC curve. The data revealed that the areas under the ROC curve (AUC) are 0.8936 (Figure 2H, P<0.0001), suggesting that CASC15 expression could be a potent candidate to identify tumor tissues from nontumorous tissues.
Overexpression Of lncRNA CASC15 Attenuated Cell Growth, Migration, And Invasion In Ovarian Cancer Cells In Vitro
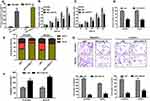
Because OVCAR3 and SKOV3 cells had the lower expression of CASC15 in the panel of ovarian cancer lines (Figure 2B), they both were selected to perform subsequent biological function assay. CASC15 expression was confirmed by qRT-PCR assay (Figure 3A, P<0.01). Furthermore, CCK-8 assay revealed that overexpression of CASC15 robustly suppressed the proliferation of ovarian cancer cells (Figure 3B and C, P<0.05). Then, soft-agar colony-formation assay demonstrated that overexpression of CASC15 could remarkably impair colony numbers of ovarian cancer cells (Figure 3D, P<0.01). In addition, cell cycle analysis showed that overexpression of CASC15 arrested cell cycle at G0/G1 stage (Figure 3E, P<0.01). Moreover, Caspase-3 ELISA assay displayed that overexpression of CASC15 augmented cell apoptosis by promoting caspase-3 activity (Figure 3F, P<0.01). Meanwhile, transwell assay elucidated that overexpression of CASC15 notably abrogated migration and invasion capability of ovarian cancer cells (Figure 3G, P<0.01).
miR-221 Was A Target Of CASC15
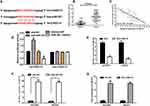
To explore whether CASC15 served as an endogenous RNA (ceRNA) or a molecular sponge to facilitate ovarian cancer development and progression, we used bioinformatics tools (Starbase V2.0 and miRcode) to find the potential targeted miRNA of CASC15. The results uncovered that miR-221 possessed complementary binding sites with CASC15 (Figure 4A). Subsequently, expression of miR-221 in ovarian cancer tissues was determined by qRT-PCR, and the result indicated that miR-221 was dramatically increased in ovarian cancer tissues (Figure 4B, P<0.01), implying that CASC15 was negatively associated with miR-221. Thus, Pearson’s correlation analysis further confirmed a negative association between miR-221 and CASC15 in ovarian cancer samples (Figure 4C, P<0.001, r=−0.7752). Subsequently, dual-luciferase reporter assay illuminated that miR-221 mimics could apparently repress the luciferase activity of the wt-CASC15 and miR-221 inhibitor (anti-miR-221) could dramatically intensify the luciferase activity of wt-CASC15, while not observed in that of mut-CASC15 (Figure 4D), suggesting that CASC15 directly interacted with miR-221. Moreover, we observed that CASC15 overexpression significantly diminished the expression of miR-221 in ovarian cancer cells (Figure 4E, P<0.01). On the other hand, the expression of CASC15 was markedly upregulated in the miR-221 underexpressed ovarian cancer cells (Figure 4F, P<0.01), suggesting that there was a reciprocal repression between CASC15 and miR-221. Subsequently, the biotin-labeled pulldown system was applied to further confirm whether CASC15 could pull down miR-221 (Figure 4G, P<0.01). We observed a large amount of miR-221 in ovarian cancer cells pulled down by biotinylated CASC15. Taken together, these results showed that miR-221 could directly bind to CASC15 at the miRNA recognition site.
ARID1A Was A Novel Target Of miR-221 In Ovarian Cancer Cells
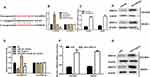
To search candidate target of miR-221, bioinformatics online tool (Starbase v2.0) predicted that miR-221 possessed the matched binding site with ARID1A (Figure 5A). Luciferase reporter assay demonstrated that overexpression of miR-221 significantly inhibited ARID1A 3ʹ-UTR activity but had no effect on the mutant ARID1A 3ʹ-UTR activity, while downregulation of miR-221 (anti-miR-221) obviously promoted ARID1A 3ʹ-UTR activity and had no effect on the mutant ARID1A 3ʹ-UTR activity (Figure 5B). Furthermore, downregulation of miR-221 strikingly enhanced the ARID1A mRNA and protein expression in ovarian cancer cells (Figure 5C and D). To explore whether CASC15 competitively inhibited the binding of miR-221 to ARID1A, we performed luciferase reporter assays. The data demonstrated that CASC15 could counteract the inhibitory effect of miR-221 on ARID1A 3ʹ-UTR activity (Figure 5E). In addition, overexpression of CASC15 significantly augmented mRNA and protein levels of ARID1A in ovarian cancer cells (Figure 5F and G). Thus, we concluded that ARID1A was a direct target of miR-221 and positively modulated by CASC15 in ovarian cancer cells.
miR-221/ARID1A Mediated Suppressive Roles Of CASC15 On Cell Viability, Apoptosis, Migration, And Invasion On Ovarian Cancer Cells
Subsequently, we validated whether CASC15 could exert its suppressive effects on cell growth, migration, and invasion through CASC15/miR-221/ARID1A axis. Notably, miR-221 overexpression decreased the expression of ARID1A in CASC2-overexpressing ovarian cancer cells (Figure 6A–C P<0.01). Functionally, miR-221 overexpression could reduce the promoted effect of CASC15 on growth of ovarian cancer cells (Figure 6D–F, P<0.05) by CCK-8 assay and soft-agar colony-formation assay. Besides, miR-221 overexpression could abrogate the promotion of apoptotic cells in CASC15-overexpressing ovarian cancer cells (Figure 6G, P<0.05). Additionally, miR-221 overexpression could decrease inhibitory effect of migration and invasion in CASC15-overexpressing ovarian cancer cells (Figure 6H and I, P<0.05). Therefore, our results showed that CASC15 could exert its inhibitory effects on the growth and migration of ovarian cancer via CASC15/miR-221/ARID1A axis.
Discussion
Recently, multiple studies have reported that lncRNA CASC15 exerts its oncogenic role in many tumors. For instance, lncRNA CASC15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop.11 LncRNA CASC15 regulates gastric cancer cell proliferation, migration, and epithelial–mesenchymal transition by targeting CDKN1A and ZEB1.12 LncRNA CASC15 is upregulated in hepatocellular carcinoma and facilitates hepatocarcinogenesis.15 However, the roles and underlying mechanisms of CASC15 remains elusive in ovarian cancer cell.
In our report, we for the first time showed that lncRNA CASC15 was strongly underexpressed, and CASC15 expression could be a poor prognostic biomarker of ovarian cancer patients. As revealed by functional experiments, CASC15 overexpression depressed biological function of ovarian cancer cells. These data evidently clarified that CASC15 acted as a tumor-suppressive gene in ovarian cancer tumorigenesis. Our conclusion was consistent with previous studies that CASC15 served as a tumor suppressor in neuroblastoma and acute leukemia.27,28
Mounting evidence has demonstrated that lncRNAs act as ceRNAs for miRNAs to modulate tumorigenesis.7 To further explore the underlying mechanism of CASC15 in the ovarian cancer tumorigenesis, we performed bioinformatics analysis indicating that miR-221 could be one target of CASC15. Intriguingly, a negative relationship between CASC15 and miR-221 was observed. Subsequently, luciferase reporter assays further manifested that CASC15 possessed the miR-221 binding site. Above all, a reciprocal repression of CASC15 and miR-221 was observed in ovarian cancer cells. So, CASC15 exerted a tumor-suppressive effect on ovarian cancer by negatively modulating miR-221 expression. Meanwhile, it was reported that overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer and miR-221 regulates proliferation and apoptosis of ovarian cancer cells by targeting BMF, suggested that miR-221 exerts oncogenic functions in ovarian cancer.18,19 Because miR-221 and miR-222 are clustered genes, which share same seed sequences and have some overlapping functions.29 In addition, Sun found that miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27kip1.30 And miR-221 and miR-222 were closely associated with the expression of CDKN1B, CDKNIC, and OS of ovarian cancer patients.31 So we speculated that there is also one interaction of CASC15/miR-222 and should be confirmed in our future study.
To further examine the potential downstream targets of miR-221, bioinformatics tools were used and the results elucidated that ARID1A could be a downstream target of miR-221. In previous studies, ARID1A has been well studied in some kind of malignancies and was found to be a tumor suppressor in ovarian cancer. Consistent with our findings, Yang et al reported that miR-221 and miR-222 simultaneously target ARID1A and enhance proliferation and invasion of cervical cancer cells.32 Subsequently, our results revealed that CASC15 could negatively modulate the expression of ARID1A via targeting miR-221 in ovarian cancer cells. Moreover, miR-221 mediated the tumor-suppressive role of CASC15 in ovarian cancer cells. Altogether, these data totally illuminated that CASC15 could abrogate growth and migration via CASC15/miR-221/ARID1A axis in ovarian cancer. Additionally, the subcellular location of CASC15, miR-221, and ARID1A deserves further investigation as the location might affect the function and structure of lncRNA-miRNA-target axis. Yin et al and Wu et al reported that CASC15 was distributed both in nucleus and cytoplasm of melanoma and gastric cancer cells, implying that CASC15 may exert regulatory function at transcriptional and post-transcriptional levels.12,16 Moreover, subcellular fractionation experiments revealed that CASC15 is predominantly a nuclear-localized lncRNA in all B-ALL cell lines tested.27 So we would further investigate the location to validate this interaction in our future study.
In short, our data firstly unveiled that CASC15 was downregulated and associated with poor prognosis of ovarian cancer patients. And CASC15 decreased ARID1A expression by sponging miR-221 to exert as one suppressive role in the development and progression of ovarian cancer. These data fully clarified that CASC15 could act as a tumor-suppressive gene and be a potential therapeutic target for ovarian cancer treatment.
Abbreviations
lncRNAs, long noncoding RNAs; CASC15, cancer susceptibility candidate 15; qRT-PCR, quantitative real-time polymerase chain reaction; OS, overall survival; PFS, progression-free survival.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author Contributions
Yin Shi and Shanshan Gao conceived and designed this project; Fan Ruan wrote the manuscript; Yin Shi and Ying Zheng conducted the experiments; Ying Zheng and Mukun Yao collected the samples and analyzed all the data. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Eisenhauer EA. Real-world evidence in the treatment of ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii61–viii65. doi:10.1093/annonc/mdx443
3. Kaldawy A, Segev Y, Lavie O, Auslender R, Sopik V, Narod SA. Low-grade serous ovarian cancer: a review. Gynecol Oncol. 2016. doi:10.1016/j.ygyno.2016.08.320
4. Morice P, Gouy S, Leary A. Mucinous ovarian carcinoma. N Engl J Med. 2019;380(13):1256–1266. doi:10.1056/NEJMra1813254
5. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. doi:10.1172/JCI84421
6. Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2017;48:8–15. doi:10.1016/j.gde.2017.10.004
7. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
8. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
9. Gao R, Fang C, Xu J, Tan H, Li P, Ma L. LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145. Arch Biochem Biophys. 2019;663:183–191. doi:10.1016/j.abb.2019.01.005
10. Shan S, Li HF, Yang XY, et al. Higher lncRNA CASC15 expression predicts poor prognosis and associates with tumor growth in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23(2):507–512. doi:10.26355/eurrev_201901_16862
11. Yu L, Xu Q, Yu W, Duan J, Dai G. LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop. Biochem Biophys Res Commun. 2018;506(4):819–825. doi:10.1016/j.bbrc.2018.10.131
12. Wu Q, Xiang S, Ma J, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12(6):799–813. doi:10.1002/1878-0261.12187
13. Yao XM, Tang JH, Zhu H, Jing Y. High expression of lncRNA CASC15 is a risk factor for gastric cancer prognosis and promote the proliferation of gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(24):5661–5667. doi:10.26355/eurrev_201712_14010
14. Zuo Z, Ma L, Gong Z, Xue L, Wang Q. Long non-coding RNA CASC15 promotes tongue squamous carcinoma progression through targeting miR-33a-5p. Environ Sci Pollut Res Int. 2018;25(22):22205–22212. doi:10.1007/s11356-018-2300-z
15. He T, Zhang L, Kong Y, et al. Long non-coding RNA CASC15 is upregulated in hepatocellular carcinoma and facilitates hepatocarcinogenesis. Int J Oncol. 2017;51(6):1722–1730. doi:10.3892/ijo.2017.4175
16. Yin Y, Zhao B, Li D, Yin G. Long non-coding RNA CASC15 promotes melanoma progression by epigenetically regulating PDCD4. Cell Biosci. 2018;8:42. doi:10.1186/s13578-018-0240-4
17. Xing Y, Wen X, Ding X, et al. CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Mol Ther. 2017;25(5):1209–1221. doi:10.1016/j.ymthe.2017.02.016
18. Xie X, Huang Y, Chen L, Wang J. miR-221 regulates proliferation and apoptosis of ovarian cancer cells by targeting BMF. Oncol Lett. 2018;16(5):6697–6704. doi:10.3892/ol.2018.9446
19. Li J, Li Q, Huang H, et al. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. Int J Oncol. 2017;50(4): 1087–1096. doi:10.3892/ijo.2017.3898.
20. Trizzino M, Barbieri E, Petracovici A, et al. The tumor suppressor ARID1A controls global transcription via pausing of RNA polymerase II. Cell Rep. 2018;23(13):3933–3945. doi:10.1016/j.celrep.2018.05.097
21. Bommareddy A, Rule B, VanWert AL, Santha S, Dwivedi C. Alpha-Santalol, a derivative of sandalwood oil, induces apoptosis in human prostate cancer cells by causing caspase-3 activation. Phytomedicine. 2012;19(8–9):804–811. doi:10.1016/j.phymed.2012.04.003
22. Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143(1):71–81. doi:10.1007/s00432-016-2252-y
23. Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51. doi:10.1186/s13046-017-0519-z
24. Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi:10.1016/j.canlet.2016.12.005
25. Wang Y, Liu Z, Yao B, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16(1):123. doi:10.1186/s12943-017-0702-z
26. Pan J, Li X, Wu W, et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett. 2016;382(1):64–76. doi:10.1016/j.canlet.2016.08.015
27. Fernando TR, Contreras JR, Zampini M, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16(1):126. doi:10.1186/s12943-017-0692-x
28. Russell MR, Penikis A, Oldridge DA, et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015;75(15):3155–3166. doi:10.1158/0008-5472.CAN-14-3613
29. He S, Lai R, Chen D, et al. Downregulation of miR-221 inhibits cell migration and invasion through targeting methyl-CpG binding domain protein 2 in human oral squamous cell carcinoma cells. Biomed Res Int. 2015;2015:751672. doi:10.1155/2015/751672
30. Sun C, Li N, Zhou B, et al. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27(kip1.). Oncol Lett. 2013;6(2):507–512. doi:10.3892/ol.2013.1393
31. Wurz K, Garcia RL, Goff BA, et al. MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer. 2010;49(7):577–584. doi:10.1002/gcc.20768
32. Yang Y, Zhao X, Li HX. MiR-221 and miR-222 simultaneously target ARID1A and enhance proliferation and invasion of cervical cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(8):1509–1515.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.