Back to Journals » International Journal of Nanomedicine » Volume 16
Lipid in Chips: A Brief Review of Liposomes Formation by Microfluidics
Received 29 July 2021
Accepted for publication 8 October 2021
Published 3 November 2021 Volume 2021:16 Pages 7391—7416
DOI https://doi.org/10.2147/IJN.S331639
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Mian Wang
Guo Zhang, Jiaming Sun
Department of Plastic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China
Correspondence: Jiaming Sun Email [email protected]
Abstract: Liposomes are ubiquitous tools in biomedical applications, such as drug delivery, membrane science and artificial cell. Micro- and nanofabrication techniques have revolutionized the preparation of liposomes on the microscale. State-of-the-art liposomal formation on microfluidic chips and its associated applications are introduced in this review. We attempt to provide a reference for liposomal researchers by comparing various microfluidic techniques for liposomes formation.
Keywords: liposomes, lipid vesicles, microfluidics, chips
Introduction
Liposomes, or lipid vesicles, are artificially synthesized by one or several phospholipid bilayers entrapping an aqueous core, sizing from nanometers to hundreds of micrometers. First discovered in the 1960s,1,2 liposomes have become a practical tool in drug delivery,3–6 membrane science7–9 or synthesis of artificial cells10–12 due to their similarity to cells and natural vesicles and simplicity to form, operate and modify.
The overwhelming success of lipid-based mRNA vaccines has brought a glimpse of hope in addressing the COVID-19 pandemic.13,14 Being a safe and efficacious carrier, liposomes can protect loaded materials from external degradation and thus are being rapidly developed as a multifunctional vaccine adjuvant-delivery system with a high capability in inducing desired immune responses for cancer treatment and vaccines.15–18 Moreover, breakthroughs in lipid-mediated gene delivery techniques such as targeted genome editing have been reported recently,19,20 which have ushered in a promising era for the treatment of genetic diseases. Meanwhile, liposomes have become an indispensable tool in membrane protein studies and synthetic organisms, helping to explore new drugs for cancer and other diseases, as well as elucidating complicated cellular functions.21–23
The prevalence of liposomes in research facilitates the search for preparation methods in an economical and feasible way. In addition to reverse evaporation24 and ethanol injection,25 commonly used production methods include thin-film hydration,26,27 extrusion,28,29 electroformation,30,31 freeze-drying32,33 and double emulsion.34,35 Though commonly utilized because of their usability, these macroscale techniques are criticized for their inability to achieve precise control over the size and polydispersity of liposomes and insufficient exploitation of reagents and materials. For example, artificial cellular compartments require precise shape and size control to emulate living cells, yet it is extremely challenging (almost impossible) to achieve such precision using macroscale techniques. It has been acknowledged that a smaller liposome size (≤100 nm) may be suitable for enhancing the activity of an antitumor drug encapsulated in liposomes.36,37 The production of “limit size” liposomes with diameters less than 50 nm is only feasible using macroscale techniques such as sonication and homogenization; however, microfluidics easily allows scalable production of SUVs with a size of 20–50 nm.38 Besides, encapsulated materials within liposomes that dissolve in aqueous solution are sometimes exceedingly expensive (such as mRNA), and bulk methods are inevitably prone to poor reproducibility and material waste. The emergence of microfluidic technology solves this predicament, as liposome formation is narrowed down to a centimeter-level chip, greatly reducing the associated cost and improving liposomes’ production efficiency and maneuverability.
As an updated version of reviews covering a similar topic published earlier,39–41 this review focuses on the current state-of-the-art liposomal preparation within microfluidic chips and related applications are introduced. A comprehensive comparative analysis of various methods classified by design rationales is conducted to highlight their advantages and disadvantages. We intend to provide insightful guidance for each method and its specific application by illustrating the strength and limitations in detail.
Characteristics of Liposome Formation
Size and Lamellarity
Lipid vesicles can be classified as unilamellar vesicles (UVs), oligolamellar vesicles (OLVs), multilamellar vesicles (MLVs) and multivesicular vesicles (MVVs) according to their lamellarity.42 Furthermore, UVs can be divided into small unilamellar vesicles (SUVs) with a particle size of <100 nm, large unilamellar vesicles (LUVs) with a particle size between 100 and 1000 nm, and giant unilamellar vesicles (GUVs) with a particle size > 1 μm according to their size (Figure 1). Generally, dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA) techniques are used to evaluate the size distribution of vesicles.43,44 Common methods used to determine the lamellarity of liposomes include nuclear magnetic resonance (NMR),45 small-angle X-ray scattering (SAXS)46 and transmission electron microscopy (TEM),47 to mention a few.
As a drug delivery system, MLVs and MVVs are more commonly used to achieve long-term drug delivery effects.48,49 To effectively and efficiently function, liposomes need to be prepared at a certain size to allow absorption into cells. Size is a critical parameter in determining liposome drug encapsulation, as well as half-life-in circulation and a smaller liposome diameter, which might be favorable in terms of optimizing in vivo drug release.50 Thus, SUVs are preferred as drug carriers for vaccines and antitumor drugs, to name a few.36,51 Since they are similar in size to cells, GUVs are more commonly used in membrane science and artificial cell synthesis.52 Differences in size result in differences in properties and applications, but monodispersity, one of the main motivators for utilizing microfluidic chips to prepare liposomes, must be ensured to maintain reproducibility.
Stability
The stability of vesicles is not only affected by the size and lipid composition but also by the physicochemical environment (such as pH and temperature) across the vesicles.53,54 For instance, interbilayer-crosslinked MLVs made by Moon et al retained ~95% of their entrapped proteins when stored in PBS at 4°C for over 30 days.55 Incorporation of pupylation has also become a popular approach used to enhance stability.56,57 Compared to polyethylene glycol (PEG)-modified liposomes that could basically be unchanged after 2 months of storage, conventional liposomes lacking PEG-surfactants showed progressive aggregation and precipitation, suggesting PEG-modification improved the vesicles’ stability.57 The surface electrostatic potential of liposomes has been shown to play a key role in the binding constant and is involved in the uptake of active compounds.58 Generally, charged liposomes are not prone to aggregation due to electrostatic repulsion.59 The Zeta potential of liposomes, used to approximate the surface charge, can be determined by laser Doppler velocimetry (LDV).60
Encapsulation Efficiency
Encapsulation efficiency (EE) is defined as the ratio of the drug encapsulated to the initial amount of drug in the system. It can be affected by many factors, such as drug properties, liposome size, and lamellarity and preparation methods. Effective separation of liposomes from the sample is the most critical step in determining EE. Commonly used separation methods for determining EE include centrifugation, dialysis, chromatography and ultrafiltration, to name a few. Quantification can be conducted after the separation and disruption of liposomes. Yet, NMR and fluorometry can directly determine EE without the separation step avoiding errors caused by the process (Table 1).
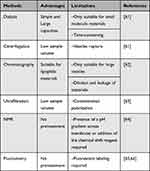 |
Table 1 Methods for Determining the Encapsulation Efficiency of Liposomes Comparison |
Yamamoto et al developed nanoparticle exclusion chromatography for determining EE without pretreatment. Only 5 μL of a liposomal suspension and 3 min of analysis time were required to quantify the amount of unencapsulated drugs.67 Variance in separation methods may lead to different results. Therefore, it is necessary to choose a suitable method to obtain reliable EE. Notably, high EE does not imply better performance. Bioactive molecules such as proteins may be compromised by various physicochemical factors during the encapsulation, resulting in only a few liposomes that ultimately function normally.39
Microfluidic-Assisted Formation of Liposomes
Hydrodynamic Flow Focusing
This method was first proposed by Jahn et al to prepare nanoscale lipid vesicles in microfluidic chips (Figure 2).68 The central channel of the lipids dissolved in alcohol was vertically connected to the two side channels with aqueous solutions. As the alcohol was mixed and diluted by the aqueous solutions to a critical concentration, the lipids self-assembled into liposomes spontaneously. Hydrodynamic flow focusing (HFF) can prepare monodisperse liposomes, and their size can be tuned by adjusting the flow rate ratio (FRR) of the water phase to the lipid phase. Subsequent research confirmed that liposomes with similar size distribution could be prepared by adjusting FRR with different chip geometries and a decrease in FRR increases the vesicle size.69–71 The lipids composition and concentration are also identified as the primary factors influencing the size of the liposomes and polydispersity index.72,73 To further study the self-assembly processes of liposomes, Jahn et al further combined an HFF device with propane jet-freezing. Disclike intermediate structures formed at the interface of lipid−alcohol and buffer, revealing more details about the formation of nonequilibrium liposomes.74
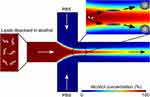 |
Figure 2 Schematic of an example of hydrodynamic flow focusing (HFF) method. Schematic of the liposome formation process by HFF. Color contours present the concentration ratios of alcohol to aqueous buffer. Reprinted with permission from Jahn A, Lucas F, Wepf RA, Dittrich PS. Freezing continuous-flow self-assembly in a microfluidic device: toward imaging of liposome formation. Langmuir. 2013;29(5):1717–1723. doi:10.1021/la303675g. Copyright 2013 American Chemical Society.74 |
Balbino et al designed a double hydrodynamic focusing (DHF) device by adding an additional pair of channels based on HFF to produce unilamellar cationic liposomes.75 DHF doubled the production rate compared to HFF and can also adjust FRR to form liposomes possessing a smaller diameter, while the polydispersity index (PDI) was no different from HFF.71,76
To further increase the production rate, Hood et al fabricated a three-dimensional microfluidic hydrodynamic focusing (3D-MHF) device using a concentric capillary array consisting of seven identical glass capillaries collectively fused in a circular pattern.77 Poly(ether-ether-ketone) (PEEK) tubings served as the lipid feed line and were threaded through the center of the array (Figure 3A). The 3D-MHF device enabled the synthesis of nanoscale liposomes with low PDI at a production rate of four orders of magnitude higher than previous HFF methods due to the entirely radially symmetric diffusion of ethanol-lipid solution. FRR and intra-annular capillary orifice size play important roles in the size control of liposomes prepared by 3D-MHF. However, the usability was compromised due to the complex assembly process of capillary arrays.
 |
Figure 3 Schematic of modified HFF devices. (A) An example of 3D microfluidic hydrodynamic focusing (3D-MHF). This device consists of a narrow bore capillary tubing surrounded by a glass multicapillary array. Liposomes are prepared as an alcohol-solubilized lipid solution continuously injected into the intra-annular capillary tubing and hydrodynamically focused in three-dimensions by a flow of aqueous buffer. Not to scale. Republished with permission of Royal Society of Chemistry from Hood RR, DeVoe DL, Atencia J, Vreeland WN, Omiatek DM. A facile route to the synthesis of monodisperse nanoscale liposomes using 3D microfluidic hydrodynamic focusing in a concentric capillary array. Lab Chip. 2014;14(14):2403–2409. Permission conveyed through the copyright clearance center, Inc.77 (B) Vertical flow focusing (VFF) technique. In the VFF design (not to scale), the focusing axis is perpendicular to that of a conventional MHF device (top). Photographs of a VFF device with a focusing channel aspect ratio of 100:1 (bottom). Reproduced Hood RR, DeVoe DL. High-throughput continuous flow production of nanoscale liposomes by microfluidic vertical flow focusing. Small. 2015;11(43):5790–5799. doi:10.1002/smll.201501345. With permission from © 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.78 |
Soon after, a vertical flow focusing (VFF) device was introduced by the same team. This device was made of a precisely aligned multilayer thermoplastic device with surprisingly low aspect ratios of 1:100 (Figure 3B). Monodisperse liposomes (80 to 200 nm in diameter) were prepared at production rates as high as 95 mg/h using this method.78 Based on the VFF design, Michelon et al and Chen et al used soft lithography or 3D printing to fabricate similar microfluidic chips for high-throughput synthesis of liposomes, respectively.79,80 Although using PMDS to fabricate VFF is a clever alternative to enhance the throughput of liposome synthesis, patterning microscale features becomes increasingly difficult as the aspect ratio increases in conventional planar microfabrication processes like soft lithography. Therefore, the throughput of liposomes was curbed as maximum aspect ratios were constrained to 6:1 in the study by Michelon et al.79 The VFF device manufactured by Hood et al overcomes these limitations and is more impressive and practical. Meanwhile, Chen et al successfully fabricated VFF chips using stereolithography, but the minimal widths of the channel were only at 200 μm due to the limited resolution of the commercial printer.80 Still, it is believed that 3D printing technologies offer great potential for realizing future improvements in tunable and high-throughput synthesis of nanoscale liposomes.
Considerable efforts have been made to utilize HFF design to generate liposomes for drug delivery. For example, liposomes encapsulating plasmid DNA synthesized by HFF could efficiently deliver genes into HeLa cells.81 Ran et al successfully prepared liposomes of various formulations by HFF, which were then further studied for physicochemical properties.82 Although high-throughput HFF-based methods like VFF can perform rapid and robust large-scale preparations of liposomes in chips, they suffer from unavoidable low EE. It was reported that EE of both methotrexate-loaded liposomes prepared by DHF or β-carotene-loaded liposomes prepared by VFF were around 60%.79,83 Hood et al integrated purification and remote drug loading elements into the HFF device. Though this device reduced the total on-chip residence time of drug-loaded liposomes to 3 minutes unprecedentedly, EE was approximately 72%, which was less compared to conventional protracted remote loading (>95%).84,85 Since the formation of vesicles is determined by mixing and diluting solvent into the aqueous solution, solvents such as ethanol cannot be completely removed from liposomes, which might affect the stability of the membranes. Last but not least, HFF-based devices, while practical, show limitations in terms of the sizes and amounts of material generated. For instance, liposomes with diameters less than 50 nm produced by HFF are only achieved at flow rate ratios of 30 or higher, resulting in substantial material dilutions.70 The emergence of a microfluidic staggered herringbone mixer (SHM) could solve the above problem.
Micromixer
Similar to HFF, liposomes can also be formed by using a microfluidic micromixer. As a powerful and general approach to generate liposomes with smaller sizes, SHM designed by Stroock et al was used to mix streams of flows effectively in microchannels. The special staggered herringbone patterns on the channel floor greatly improve mixing efficiency, which can be altered by the asymmetry of the herringbones and the number of herringbones per half cycle.86 Zhigaltsev et al pioneered SHM (Figure 4A) as a scalable manufacturing method to produce limit size lipid nanoparticles (LNPs) as small as 20 nm by millisecond mixing of lipids dissolved in ethanol with an aqueous stream. They demonstrated the excellent ability of nanoscale liposomes synthesized by SHM to load and retain doxorubicin and confirmed the bilayer structure of SUVs by 31P NMR and cryogenic TEM (cryo-TEM).87 They further reported that by using SHM, limit size SUVs with a diameter of around 20 nm could be produced at FRR of only 3 (1.5 mL/min for the flow rates of the aqueous channel).
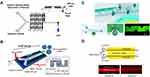 |
Figure 4 Lipid nanoparticles formulation process employing micromixers. (A) Schematic of lipid nanoparticles formulation process employing the staggered herringbone microfluidic mixing device. Reprinted with permission Zhigaltsev IV, Belliveau N, Hafez I, et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir. 2012;28(7):3633–3640. doi:10.1021/la204833h. Copyright 2012 American Chemical Society.87 (B) Invasive lipid nanoparticle production (iLiNP) device. Three-dimensional and top views of the iLiNP device with the basic structure of 20 baffle mixer structure sets. Reproduced with permission of American Chemical Society from Kimura N, Maeki M, Sato Y, et al. Development of the iLiNP device: fine tuning the lipid nanoparticle size within 10 nm for drug delivery. ACS Omega. 2018;3(5):5044–5051. https://pubs.acs.org/doi/10.1021/acsomega.8b00341. Further permissions related to the material excerpted should be directed to the ACS.95 (C) The acoustically driven microfluidic micromixer. Fluorescence suggested the mixture of lipid solvents and water in the presence of an acoustic field. Republished with permission of Royal Society of Chemistry from Rasouli MR, Tabrizian M. An ultra-rapid acoustic micromixer for synthesis of organic nanoparticles. Lab Chip. 2019;19(19):3316–3325. Permission conveyed through the copyright clearance center, Inc.96 (D) Schematic illustration of the electrohydrodynamic micromixer. Fluorescence indicated the laminar streams of water and ethanol when the voltage is off and on. Reprinted with permission from Modarres P, Tabrizian M. Electrohydrodynamic-driven micromixing for the synthesis of highly monodisperse nanoscale liposomes. ACS Appl Nano Mater. 2020;3(5):4000–4013. doi:10.1021/acsanm.9b02407. Copyright 2020 American Chemical Society.97 |
Kastner et al found that the increase of FRR results in increased PDI but decreased size of liposomes prepared by SHM.88 However, the results of Maeki et al showed that the size and PDI of liposomes prepared by SHM decreased with the increase of FRR simultaneously.89 Besides, the SHM cycle numbers and the position of the first SHM are also significant factors for the production of vesicles. For example, the size of the liposomes gradually increased with decreasing the SHM cycle numbers and increased distance from where the first SHM was located. Due to insufficient mixing of the aqueous and lipid solutions observed under the high FRR condition, Maeki et al assumed that rather than complete mixing of the solutions, the rapid dilution of ethanol to a critical concentration was essential to form vesicles.89 In addition to the parameters of SHM itself, the lipid composition,90 concentration91 and even solvent selection92 are all considered key parameters in liposomes formation. It should be pointed out that not all LNPs prepared by SHM constitute a continuous bilayer that qualifies them as liposomes; some LNPs are micelle-like structures (micelles) with a non-aqueous core.93 Unfortunately, techniques such as cryo-TEM were unaffordable or just unreported in some studies to confirm the structure of LNPs prepared by micromixer. According to those reported results, the formation of liposomes or micelles in SHM might depend mainly on lipid composition and synthesis parameters.87,90,92,94
Instead of SHM, Kimura et al designed an invasive lipid nanoparticle production (iLiNP) device consisting of 20 sets of zigzag-shaped mixer structures (Figure 4B). iLiNP devices enabled the LNPs size tuning at 10 nm intervals in size ranging from 20 to 100 nm precisely by various baffle parameters, flow rate and FRR, which could not be achieved by conventional SHM.95 Notably, encapsulated materials such as mRNA and siRNA dissolved in aqueous solution are expensive so the LNP must be produced at low and constant FRR conditions. iLiNP device could produce LNPs in size ranging 20–40 nm at flow rates of 50, 100, and 500 μL/min and an FRR of 9.
To further enhance the speed and homogeneity of the mixing, Rasouli et al introduced an acoustically driven microfluidic micromixer (Figure 4C).96 The acoustic microstreaming reinforced by integrating sharp edges and bubbles in this device triggered higher throughput of liposomes with lower PDI. Compared to passive micromixers, the intense and pervasive acoustic micro-streams in this device prevented the formation of large nanoparticle aggregates and clogging of the channel. However, the vibrational amplitude of sharp edges rapidly diminished as the flow rate increased, limiting the throughput. Besides, the fabrication of the whole device was somehow complicated for non-professionals; thus, the usability was compromised.
Subsequently, the same team applied an electrohydrodynamic micromixer to prepare liposomes with a narrower size distribution at a throughput of 1∼2 × 1010 min−1 (Figure 4D).97 The liposome size was determined mainly by the initial lipid concentration and the FRR in this device. Compared to conventional micromixers that require soft lithography to fabricate grooves, this novel device is easier to implement with only an additional metal deposition step. It is worth mentioning that although the electric field can avoid contaminant clogging or liquid leakage, liposome electroporation, as well as lipid oxidation may occur, affecting the size and stability of liposomes and even the passive permeability of the lipid bilayers.98,99
SHM can apparently produce high quantities of limit size liposome/LNPs at lower FRR, avoiding substantial material dilutions and waste. However, problems with relatively low EE have been reported. In the studies by Joshi et al, EE of liposomes prepared by SHM loading a hydrophilic drug (metformin) or a lipophilic drug (glipizide) was around 20% or 40%, respectively. When used to encapsulate highly lipophilic propofol, EE was approximately 40% which is significantly higher than EE in liposomes prepared via sonication (15%).100 It is worth mentioning that LNPs with micellar structures produced by SHM had higher EE when encapsulating biological molecules such as siRNA.101 Belliveau et al used a device with six parallel SHM elements to produce limit size LNPs composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/cholesterol to encapsulate siRNA at an extraordinary throughput (72 mL/min or 580 mg LNPs/min), while EE of siRNA was in excess of 95% and PDI was below 0.1.102 These LNPs exhibited an electron-dense core in cryo-TEM, suggesting micellar structures. Such morphology was distinctly different from bilayer LNPs, which exhibited an electron-dense ring and less dense interior associated with unilamellar vesicle systems with an aqueous interior. It was further proved by Leung et al that high siRNA encapsulation levels for siRNA in LNP systems were inconsistent with bilayer structure since encapsulation was dependent on the presence of cationic lipid.103
Generally, to prepare liposome/LNPs for drug delivery, micromixers may be a better choice because of their relatively high EE, easy usability and full use of encapsulated materials. The throughput of micromixers had been exponentially scaled up by incorporating an array of numerous mixing channels that operate simultaneously.104 In fact, commercial devices based on micromixers such as NanoAssemblr™ platforms have been universally used in many studies to prepare liposome/LNPs for different applications, such as CRISPR-Cas9 genome editing for cancer therapy105 and in utero mRNA delivery for monogenic fetal diseases.106 Notably, COVID-19 vaccine nanoparticles manufactured by Pfizer are reported to be prepared by microfluidic mixers.93
Emulsion Templates
Unlike HFF and micromixers, discrete droplets with defined sizes generated by immiscible fluids serve as templates for the formation of liposomes in this method. Generally, these liposomes with diameters measured in micrometers are more used in membrane science and artificial cell research in contrast to drug delivery.
Single Emulsion Templates
Weiss et al developed an approach that merges vesicles and water-in-oil (W/O) droplets to generate cell-like compartments.107 In this method, LUVs prepared by extrusion were first encapsulated in copolymer-stabilized W/O droplets. By introducing Mg2+solution during droplet production, a contiguous lipid bilayer at the inner interface of the droplet was formed due to the transformation of the encapsulated LUVs, which they named droplet-stabilized GUVs (dsGUVs). Assembled lipid compartments could be released from the droplets into the aqueous phase by another microfluidic device (Figure 5). Before demulsification, various proteins and molecules could be loaded sequentially into dsGUVs using pico-injection technology (inspired by Weitz lab designs)108 to form synthetic cells. By combining different microfluidic technologies, copolymer-stabilized W/O droplets were used as templates for high-throughput preparation of GUVs (103/s) with unique and precise compositions. This elegant technique ensured high EE, and no traces of remaining oil could be detected in the released GUVs under a Raman microscope. A bottom-up reconstitution of the actin cytoskeleton, microtubules or even FoF1-ATP synthase within dsGUVs was successfully obtained by this technology, overcoming fundamental limitations for the design of complex synthetic cells.
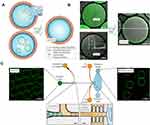 |
Figure 5 Formation droplet-stabilized GUVs. (A) LUVs or GUVs were encapsulated in W/O copolymer-stabilized droplets by microfluidics. To transform the encapsulated vesicles into a supported lipid bilayer at the copolymer-stabilized droplets’ inner interface, 10mM Mg2+ was applied during droplet production. (B) Representative combined images of green fluorescence from lipids and bright-field microscopy of the encapsulated LUVs and GUVs, and of the dsGUVs. Scale bar, 10 μm. (C) The microfluidic device was designed to release the assembled lipid compartments from the surrounding stabilizing polymer droplets into the aqueous phase. The left image shows the fluorescence images of the monodisperse dsGUVs in the oil phase prior to release. After injection, droplets are separated from each other at a T-junction where a tributary oil flow containing 20 vol% destabilizing surfactants merges with the droplet flow. Passive trapping structures enable the draining of the continuous oil phase and decelerate the droplets before they enter the aqueous phase. A comparison of dsGUVs and released lipid compartments shows that they are comparable in size. Scale bars, 20 μm. Reprinted with permission from Springer Nature Customer Service Centre GmbH: [Springer Nature] [Nature materials] Weiss M, Frohnmayer JP, Benk LT, et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat Mater. 2018;17(1):89–96. Copyright 2018. https://www.nature.com/articles/nmat5005.107 |
The same team further reduced the diameter of dsGUVs to ~2 μm by a microfluidic droplet splitter, paving the way towards a targeted delivery of advanced cargos such as microparticles, viruses or macromolecular DNA-robots.109 They investigated interactions between differently charged GUVs and cells and proved small GUVs to be a practical alternative for transporting and delivering very large, complex cargos such as baculovirus to specific cells, which is not possible by conventional SUV-based delivery. The problem was that droplet-splitted GUVs was not mechanically stable enough, thus leading to a low yield of daughter GUVs. Nonetheless, their sharp control over the GUVs composition and functionalization allowed the construction of GUVs with functional diversity and high targeting specificity. However, the shortcomings of single emulsion templates include the challenging requirement for soft lithography (the minimum width of a microchannel of droplet splitter is less than 2 μm) and the special demand of pico injection devices.
Ice Droplet Hydration
First presented by Sugiura et al,110 lipid-coated ice droplet hydration method was used to prepare monodisperse W/O droplets between 4 and 20 µm in diameter as templates. After freezing, the droplets precipitated and separated, followed by the replacement of hexane solution containing lipid. Next, the solvent was evaporated, while the water droplets were still frozen, and an aqueous solution was added, resulting in the formation of oligolamellar or multilamellar giant vesicles. However, UVs must be yielded by extrusion in this method, and EE decreases from 35% to 12% after extrusion, which restrains the practicability.110 This method was applied successfully for the entrapment of an enzyme (α-chymotrypsin), but the problem of low EE was not solved since partial leakage from the initially formed droplets could not be prevented.111
Droplet Emulsion Transfer
Pautot et al first described the droplet emulsion transfer method.34,112 W/O emulsion prepared by stirring the mixture of aqueous solution and lipid suspension was again transferred to the aqueous phase. Droplets in the emulsion pulled the second monolayer from the oil–water interface to complete the bilayer, resulting in the formation of UVs (Figure 6A). They successfully encapsulated various macromolecules like G-action into GUVs as micro-bioreactors with up to 98% EE. Asymmetric GUVs with a controllable composition of each leaflet independently could also be prepared using this preparation technique. However, these GUVs were polydisperse since the emulsion was prepared by stirring. Further refinements utilizing microfluidic devices then solved the problem (Figure 6B). Monodisperse GUVs could be prepared from lipid-stabilized W/O droplets using microfluidic droplet formation techniques such as T-junction and flow focusing. However, droplets must be recollected to transfer across the oil–water interface.113–115 Importantly, it was reported by Pautot et al that only a fraction of the larger emulsion droplets would pass through the interface to form liposomes because most were likely to break at the interface.112 They also observed that asymmetric vesicles required several hours of equilibration time to achieve complete lipid coverage of the second monolayer to convert droplets into unilamellar vesicles. Unfortunately, shorter equilibration times (<2 h) led to insufficient coverage and uncontrolled lipid composition, while longer equilibration times (>3 h) resulted in an accumulation of lipid in multilayer structures, creating a crucial limitation in the technique.
 |
Figure 6 Methods using droplet emulsion transfer to prepare liposomes. (A) Droplet emulsion transfer method for vesicle formation. Reproduced from Pautot S, Frisken BJ, Weitz DA. Engineering asymmetric vesicles. Proc Natl Acad Sci. 2003;100(19):10718. doi:10.1073/pnas.1931005100. With permission of (2003) National Academy of Sciences, U.S.A.34 (B) Giant liposome formation device using W/O emulsion. Reprinted with permission from Hu PC, Li S, Malmstadt N. Microfluidic fabrication of asymmetric giant lipid vesicles. ACS Appl Mater Interfaces. 2011;3(5):1434–1440. doi:10.1021/am101191d. copyright 2011 American Chemical society.114 (C) Integrated microfluidic device by droplet transfer method for vesicle formation. Reproduced from Matosevic S, Paegel BM. Stepwise synthesis of giant unilamellar vesicles on a microfluidic assembly line. J Am Chem Soc. 2011;133(9):2798–2800. doi:10.1021/ja109137s. https://pubs.acs.org/doi/10.1021/ja109137swith permission of American Chemistry Society.116 (D) Asymmetric giant liposome formation device. Reproduced from Karamdad K, Law RV, Seddon JM, Brooks NJ, Ces O. Studying the effects of asymmetry on the bending rigidity of lipid membranes formed by microfluidics. Chem Commun. 2016;52(30):5277–5280. doi:10.1039/C5CC10307J.118 (E) Schematic of the layer-by-layer cell membrane assembly. Phospholipid-stabilized water-in-oil droplets were first formed and trapped in the static droplet array. Lipid monolayer deposition proceeded as oil/water-phase boundaries pass over the droplets, leading to the formation of a double-bilayer vesicle after three-phase exchanges. Reprinted by permission from Springer Nature Customer Service Center GmbH: Springer Nature, Nature Chemistry, Matosevic S, Paegel BM. Layer-by-layer cell membrane assembly. Nat Chem. 2013;5(11):958–963. Copyright 2013. 119 |
Successful integration of the generation and transfer of droplets into a microfluidic chip was achieved by Matosevic et al (Figure 6C).116 They used a triangular post to drive lipid-stabilized droplets to transfer through the interface resulting in the deposition of a second lipid monolayer. However, assembly yield was only 5% as droplets were easily lost due to premature contacts with the aqueous flow. EE also decreased to 83% consequently. Karamdad et al further optimized the device to enhance liposome yield and EE.117,118 The device contained a micro-step junction manufactured by double-layer photolithography, where W/O droplets were transferred across the oil–aqueous interface to construct monodisperse asymmetric vesicles at high throughput (Figure 6D). It is worth noting that double-layer photolithography requires high professional skills.
In addition, the Paegel group cleverly integrated a static array on a microfluidic chip for lipid-stabilized W/O droplets trapping.119 By introducing oils and water alternately, each oil/water boundary crossing would deposit a new monolayer of lipids on the immobilized droplets, suggesting that a phospholipid membrane could be assembled layer-by-layer on these droplets using this platform (Figure 6E). Asymmetric multi-bilayers vesicles were successfully assembled within this chip, and this was considered a new tool for studying lipid biosynthesis enzymes and transporters.
Based on droplet emulsion transfer, Morita et al developed the droplet-shooting and size-filtration (DSSF) method to prepare monodisperse GUVs (10–20 μm) with controlled asymmetric leaflets in lipid bilayers.120 Polydisperse W/O droplets were generated from the tip of a glass capillary by centrifugation, and only 10–20% droplets with the smallest diameter were transferred through the oil–water interface selectively by kinetic size filtration (Figure 7), as previously observed by Pautot et al.112 A 1 μL sample solution was sufficient to generate hundreds of liposomes by DSSF without any wasted material. However, a limited amount of solution in the capillary makes it difficult to prepare liposomes efficiently. In addition, residual oil between the leaflets was confirmed, though the authors claimed that it was low enough not to affect the physicochemical and biochemical properties.120,121
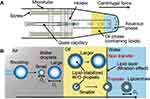 |
Figure 7 Formation of cell-sized liposomes by the droplet shooting and size-filtration (DSSF) method. (A) Capillary-based microfluidic device. (B) Generation of cell-sized liposomes (within the rectangle shown in (A)). Reproduced from Morita M, Onoe H, Yanagisawa M, et al. Droplet-shooting and size-filtration (DSSF) method for synthesis of cell-sized liposomes with controlled lipid compositions. ChemBioChem. 2015;16(14):2029–2035. doi:10.1002/cbic.201500354 with permission of Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim120. |
The main superiority of the droplets emulsion transfer method based on microfluidic devices is the preparation of monodisperse GUVs with asymmetry and high EE, making it suitable for membrane science as well as encapsulation of a diversity of bioactive macromolecules. Adir et al prepared GUVs by the emulsion transfer method and constructed a cell-free expression system by encapsulating bacterial lysate inside GUVs. The obtained synthetic cells were then used to investigate protein/RNA production and activity in an isolated environment.122 However, the problem of equilibration time, as discussed above, is still unsolved, and controversies arise over the presence of oil residues between leaflets of liposomes since droplets are transferred from oil to the aqueous phase. Kubatta et al demonstrated that an oil layer was accumulated at the top of the giant vesicles (3–5 mm) prepared by bulk transfer method, and oil remnants were also observed in the bilayer by Hu et al114,123 Yet the results of the study by Matosevic et al and Pautot et al disagreed since they both detected no trace of oil the bilayer by a thin-layer chromatography analysis or quantitative chemical probing, respectively. Inhibition of membrane protein insertion and function was also not observed.112,119 Squalene was recommended by Pautot et al due to a good continuous phase of emulsion and is immiscible in lipid bilayers.112 Nevertheless, the possibility of oil residues cannot be excluded due to the sensitivity limit of the techniques they used. It seems that different protocols could lead to contradictory conclusions. Imaging based on cryo-TEM or highly sensitive techniques, such as Raman microscopy, is required to verify membrane morphology.
Double Emulsion Templates
First proposed by Shum et al, they prepared lipid-stabilized water-in-oil-in-water (W/O/W) double emulsion droplets using glass microcapillary devices as templates. The inner phase consisted of an aqueous solution containing materials used for encapsulation. The middle phase was phospholipids dissolved in a mixture of toluene and chloroform, while the outer phase was poly(vinyl alcohol) (PVA) and glycerol solution (Figure 8A).124 As the solvent layer became thinner during evaporation, the phospholipids were concentrated and then forced to rearrange on the double emulsion templates, thereby forming GUVs. Although monodisperse GUV with defined size and high EE could be prepared by this method, a small amount of organic solvent remained between lipid leaflets after dialysis. To eliminate the presence of undesirable solvents, the same team used another glass capillary microfluidic device to produce W/O/W droplets with ultrathin oil shell (<1 μ m) as templates to prepare GUVs containing minimal residual solvent within their membrane (Figure 8B).125,126 This time, the inner phase was replaced by PEG and PVA solution, and the oil phase was a mixture of chloroform and hexane. Dewetting occurs since the chloroform evaporates more rapidly than the hexane. The hexane floated as an oil pocket at the top of droplets then evaporated eventually, thus forming GUVs with little residual oil.126 GUVs prepared by this device were used as a cell-free platform for synthesizing green fluorescent proteins, suggesting that PVA in the inner aqueous phase might not influence the bioactivity of liposomes.127 Using this technique, a therapeutic enzyme (Cu, Zn-Superoxide dismutase) was successfully encapsulated into liposomes to treat a mice ear edema model. An EE of 59 ± 6% and an enzyme activity of 82 ± 3% were obtained.128
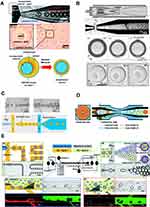 |
Figure 8 Double emulsion templates method. (A) Formation of phospholipid-stabilized W/O/W double emulsion in a glass microcapillary device. The double emulsion drops have an aqueous core surrounded by a solvent shell containing phospholipid (top). As the solvent layer becomes thinner during evaporation, the phospholipids are concentrated, forming GUVs (bottom). Reptinted with permission from Shum HC, Lee D, Yoon I, Kodger T, Weitz DA. Double emulsion templated monodisperse phospholipid vesicles. Langmuir. 2008;24(15):7651–7653. doi:10.1021/la801833a. Copyright 2008 American Chemical Society.124 (B) Preparation of water-in-oil-in-water double emulsion drops with ultrathin shells (top). The hexane floats as an oil pocket at the top of droplets then evaporate eventually, thus forming GUVs with little residual oil (bottom). Reproduced with permission of Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim from Arriaga LR, Datta SS, Kim SH, et al. Ultrathin shell double emulsion templated giant unilamellar lipid vesicles with controlled microdomain formation. Small. 2014;10(5):950–956. doi:10.1002/smll.201301904.126 (C) Schematic and microscope images of double emulsion formation in PDMS chips for liposomes preparation. Reprinted from Teh S-Y, Khnouf R, Fan H, Lee AP. Stable, biocompatible lipid vesicle generation by solvent extraction-based droplet microfluidics. Biomicrofluidics. 2011;5(4):44113–4411312. doi:10.1063/1.3665221, with the permission of AIP publishing. 129 (D) Schematic illustration of the device to produce water-in-oil-in-oil-in-water (W/O/O/W) triple emulsion droplets. Cross-section pictures correspond to the position of the dashed line in the horizontal view. Republished with permission of Royal Society of Chemistry from Arriaga LR, Huang Y, Kim S-H, et al. Single-step assembly of asymmetric vesicles. Lab Chip. 2019;19(5):749–756. doi:10.1039/C8LC00882E. Permission conveyed through the copyright clearance center, Inc.132 (E) Images of the process for forming the asymmetric vesicles in the PDMS microfluidic device. (a) Once W/O droplets were formed at the first flow-focusing region, the emulsions travel through a serpentine channel. (b and c) Images are taken at the first and the last triangular posts with fluorescent images of lipid-1 (red) with lipid-2 (green). They confirm the “dual-pinching” separation strategy for replacing lipid-1 (red) with lipid-2 (green) in the oil solution that surrounds the droplets. The excess oil/lipid-1 solution flows to the waste outlet. (d) The formation of W/O/W double emulsions (indicated by the dotted red arrow) and the excess oil droplets (indicated by the solid blue arrow) at the second flow-focusing region. Reprinted with permission from Lu L, Schertzer JW, Chiarot PR. Continuous microfluidic fabrication of synthetic asymmetric vesicles. Lab Chip. 2015;15(17):3591–3599. doi:10.1039/C5LC00520E. Copyright 2015 American Chemical Society.133 |
Double emulsion droplets were also generated in PDMS chips.129–131 Unlike the capillary microfluidic device, these W/O/W droplets comprised of an internal phase of an aqueous solution, a middle phase of oleic acid and an outer phase containing ethanol (Figure 8C). Notably, ethanol served to extract the oleic acid from the middle layer and then evaporated away at last. GUVs are formed by self-assembling of the phospholipids at the oil/water interfaces. Most GUVs prepared by this method remained stable for a long time (>3 months) and could be used for cell-free protein expression as synthetic cells.129 There might be residual oleic acid or excess phospholipid left in the lipid shell, but this method was more biocompatible than those described above since toxic solvents such as chloroform were not used. Also, this device did not require any sophisticated design principles and treatments other than an extra step of hydrophilisation of the external channel.
The double emulsion templates method can also be used to prepare asymmetric UVs. Arriaga et al further upgraded the glass microcapillary devices. Sophisticated water-in-oil-in-oil-in-water (W/O/O/W) triple emulsion droplets were used as templates for continuous high-throughput preparation of vesicles with asymmetric membranes in a single step (Figure 8D).132 Meanwhile, Lu et al. designed an integrated PDMS chip to fabricate asymmetric UVs. By their ingenious design, lipid-stabilized W/O droplets were first formed at the first flow-focusing region, then the oil/inner-leaflet-lipid solution that surrounded the droplets was replaced with the oil/outer-leaflet-lipid solution. After W/O/W droplets formed in the second flow-focusing region, asymmetric vesicles were collected off-chip followed by the extraction of oil between leaflets of the double emulsions (Figure 8E).133 The oil phase was also extracted by ethanol in this technique, indicating residual oleic acid or excess phospholipid might remain within the lipid shell. It was reported that over 80% of the liposomes remained stable for at least 6 weeks, and the membrane asymmetry was maintained for over 1 day. Compared to W/O/O/W emulsion templates, this method seems easier to handle and produces asymmetric vesicles with monodisperse sizes, high EE and high throughput. However, the choice of solvents was limited to PDMS chips. For example, a solvent typically used in glass capillary devices, chloroform would dramatically swell PDMS and create blockages in the microchannel network.134
While these results may increase the utility of vesicles with high EE as model biological membranes or for drug delivery applications, there are still some constraints in terms of usability. For instance, the preparation of liposomes by this technique requires careful handling and assembly of multiple glass capillaries or PDMS chips and precise control of several microfluidic parameters simultaneously. Five pumps were used to operate the glass capillary device at the same time in the study by Arriaga et al, not to mention the demanding selections of lipid solvents.132 Secondly, multiple components like PEG, PVA and Pluronic F-68 are also introduced into the aqueous solution during the formation, which may affect the biological properties of liposomes. Importantly, residual solvent remained after the formation of liposomes.
To resolve the oil residues in liposomes once and for all, Deshpande et al developed a microfluidic technique, namely, octanol-assisted liposome assembly (OLA), to form unilamellar, monodisperse, cell-sized GUVs with an efficient, autonomous and fast solvent-extraction mechanism (Figure 9). 135 They replaced the conventional oil phase with 1-octanol. Within a few seconds after its generation, the W/O/W droplet developed into an inner aqueous core entrapped by a lipid bilayer and an outgrowing pocket of 1-octanol containing excess lipids, which completely separated from the liposome in a few minutes. The unilamellarity of liposomes formed by OLA was confirmed by the insertion of the bilayer-spanning protein pore α-hemolysin. The non-leaky detachment of the oil phase also ensured excellent EE. Moreover, the biocompatibility of OLA-based liposomes was verified by encapsulating bacterial divisome proteins.135,136
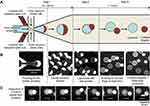 |
Figure 9 Schematic of octanol-assisted liposome assembly (OLA). (A) Schematic representation showing the working principle of on-chip production of liposomes using OLA. (B) Corresponding fluorescence images showing each of the steps described above. (C) Temporal-resolution sequences showing the separation of the 1-octanol droplet from the liposome. The first frame of the sequence in c was obtained about 1 min after the double-emulsion droplet formed. Reproduced from Deshpande S, Caspi Y, Meijering AE, Dekker C. Octanol-assisted liposome assembly on chip. Nat Commun. 2016;7:10447. doi:10.1038/ncomms10447.135 |
Similarly, Deng et al induced spontaneous and complete dewetting of W/O/W droplets to prepare oil-free monodisperse UVs using a surfactant-assisted microfluidic strategy with coaxial glass capillaries.21,137 A triblock copolymer surfactant, Pluronic F-68, was added into the outer phase to control the developing interfacial energies, prompting the oil shells to gradually dewet from the interior drops; therefore, liposomes were separated from oil pockets by excess lipids ultimately just like OLA. Floating oil droplets can be easily removed by pipetting or evaporating after injecting the mixtures into the sucrose solution. Multicompartment liposomes could be formed by varying flow rates with an additional droplet generator (Figure 10). This work is the first to realize the controllable production of multicompartment liposomes in microfluidics. However, the lipid was dissolved in a toxic organic solvent mixture of chloroform and hexane here.
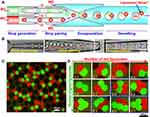 |
Figure 10 Formation of liposomes with distinct multicompartments. (A and B) Schematic and snapshots of fabrication of double emulsions with two distinct drops. (C) Confocal images of monodisperse liposomes with two different compartments. (D) As-prepared liposomes with controlled structures and various configurations. Reprinted with permission from Deng -N-N, Yelleswarapu M, Huck WTS. Monodisperse uni- and multicompartment liposomes. J Am Chem Soc. 2016;138(24):7584–7591. doi:10.1021/jacs.6b02107. Copyright 2016 American Chemical Society.137 |
Despite concerns that the external aqueous environment where liposomes are collected might be contaminated due to the rupture of oil pockets, and some 1-octanol that might remain in the bilayer, OLA addresses the limitations of other methods to a considerable degree, such as the time-consuming solvent-extraction and remnants of organic solvents in the lipid bilayer. Since the organic solvent used is bio-friendly, OLA is becoming a versatile platform for drug delivery, micro-reactors and synthetic cells. For example, the OLA platform was integrated with an optofluidic transport assay to create a complete microfluidic total analysis system for quantifying drug permeability. Norfloxacin and ciprofloxacin transport through liposomes was measured at physiological pH and salt concentrations.138 Vaezi et al produced liposomes to encapsulate taxotere using the OLA. Combined with a microfluidic cell analysis system, encapsulated liposomes’ efficacy was analyzed within a single cell in a trapping chip for quantifying drug permeability and apoptosis assay.139 The EE of taxotere-loaded liposome was 65.49 ± 3.08%, as reported.139 Niederholtmeyer et al developed porous cell-mimics capable of gene expression and communication via diffusive protein signals utilizing OLA.140 In addition, OLA has proven to be a powerful tool for controlling and studying the formation of membrane-less organelles within liposomes, which help facilitate the bottom-up creation of synthetic cells.141
Pulsed Flow Jetting
First reported by Funakoshi et al,142 this method mimicked the process of blowing soap bubbles. The pulsed jet flow generated from a micro-nozzle was applied to a planar lipid bilayer formed by the droplet contact method, as the bilayer then stretched and formed free vesicles eventually (Figure 11A).143 The size of the vesicles produced could be tuned by different jet dispensing times. A large amount of monodisperse GUVs (300 μm −600 μm) can be prepared by repeating this process, accompanied by the formation of a small amount of smaller “satellite vesicles“. However, residual organic solvent (n-decane) was observed inside the membrane. Subsequently, the same team proved that smaller “satellite vesicles” are generated due to the inhomogeneous break-up of the deformed lipid microtubes. Based on the ”satellite vesicles”, they prepared asymmetric GUVs with a diameter of 3–20 μm by modifying parameters like the area of the planar lipid bilayer and the jet dispensing time (Figure 11B).144 They confirmed little residual amount of oil in these GUVs using a Raman scattering microscope. Flip-flop motions of lipid molecules in the asymmetric GUVs were observed, indicating that the presence of n-decane between the leaflets had little effect on membrane dynamics.144 Recently, the team successfully produced cell-sized vesicles containing vesicles, namely MVV, generated by applying pulsed jet flow against two parallel planar asymmetric lipid bilayers formed in a triple-well device, similar to the formation of W/O/W droplets to some extent.145 Moreover, by applying pulsed-jet flows of longer duration and higher pressure to an asymmetric planar lipid bilayer, nano-sized asymmetric lipid vesicles were formed. The resulting vesicles, however, were polydisperse, and around 30% were multilamellar.146
 |
Figure 11 Examples of pulsed jet flow method. (A) Conceptual diagram of the “blowing vesicle” method. The green area represents organic solvent. Sequential images of vesicle formation captured by a high-speed CCD camera. The planar membrane was stretched to form a column and broken up into spherical vesicles within 10 ms. Reprinted with permission from Funakoshi K, Suzuki H, Takeuchi S. Formation of giant lipid vesicle like compartments from a planar lipid membrane by a pulsed jet flow. J Am Chem Soc. 2007;129(42):12608–12609. doi:10.1021/ja074029f. Copyright 2007 American Chemical Society.142 (B) Formation of a lipid microtube from the asymmetric planar lipid bilayer by the jet-flow method. Reproduced from Kamiya K, Kawano R, Osaki T, Akiyoshi K, Takeuchi S. Cell-sized asymmetric lipid vesicles facilitate the investigation of asymmetric membranes. Nat Chem. 2016;8(9):881–889. doi:10.1038/nchem.2537 with permission with permission of Springer Nature.144 (C) Schematic of the acoustic jetting method. The focused flow generated by the ultrasonic transducer deforms a planar lipid bilayer to form GUVs and time-series images of GUV formation. Reprinted from Armstrong M, Vahey MD, Hunt TP, Fletcher DA. Forming and loading giant unilamellar vesicles with acoustic jetting. Biomicrofluidics. 2020;14(6):064105. doi:10.1063/5.0021742, with permission from AIP publishing.151 |
Unlike the microdispenser applied by Funakoshi et al, Stachowiak et al used a piezoelectric inkjet nozzle to perform microfluidic jetting.147,148 GUVs 10–400 μm in diameter were formed by varying the pulse amplitude, the number of pulses and fluid viscosity. Utilizing this technology, they synthesized asymmetric GUVs with oriented membrane protein incorporation and constructed a synthetic system in which membrane proteins were delivered to the outside of GUVs mimicking aspects of exocytosis.149
The problem with this method is that the formation of the bilayer produced by manual pipetting requires a certain incubation time, the rate of membrane regrowth limits the vesicle production rate. Also, the difficulty in repositioning the micro-nozzle relative to the planar lipid bilayer can hinder repeatability and throughput. As a result, fast and continuous preparation of liposomes could not be prepared in large quantities via this method. The piezoelectric inkjet technology could increase the production rate by up to 200 Hz in a way, but a pause of 2–4 seconds was needed for the lipid bilayer to regrow.148 To address this problem, Gotanda et al arranged the six wells on a circular rotation platform to prepare various asymmetric liposomes sequentially by accurately controlling rotation speed parameters and jetting timing through an automated system. Approximately 300 asymmetric cell-sized GUVs (mean diameter of 8.9 ± 6.7 μm) were collected within 2 minutes.150 Armstrong et al pioneered the use of focused high-intensity ultrasound instead of micro-nozzles to induce focused flow jets to prepare GUVs (Figure 11C), eliminating stability challenges. GUVs of 100–300 μm in diameter were formed by controlling the numerical aperture of the acoustic lens and the acoustic wavelength.151 Notably, ultrasound itself will affect the stability of liposomes. It is crucial to ensure the stability of planar lipid bilayers during the formation of vesicles.
In this method, materials such as DNA, cells, and so on, regardless of their size, concentration, or chemical properties, can be directly encapsulated into liposomes by jetting, similar to pico injection, bringing about a high EE (~90%) as reported.144 However, organic solvent remnants were found between leaflets in this method.142,144,152 Thus, sophisticated equipment demanding operating conditions with low liposome yield confines its further applications other than membrane science and synthetic cells. Nevertheless, asymmetric GUVs were stably produced by Kamiya et al using pulsed flow jetting.144 These asymmetric GUVs successfully emulated a lipid flip-flop corresponding to that of apoptotic cells and showed the flop dynamics influenced by an antibiotic peptide.
Solvent-Free Method
Hydration on Chips
Thin-film hydration is a common method used for the formation of vesicles. The process involves depositing lipid films onto a substrate after evaporation of the organic solvent. The films then swell, peel off, and eventually self-assemble into liposomes in the absence of an aqueous solution. Since there is no control over the hydration process, most of the liposomes prepared are polydisperse MLVs. Subsequent extrusion or other treatments are required to obtain smaller monodisperse UVs.153–155
To handle this problem, Osaki et al focused on the control of lipid deposition process. Arrayed micro apertures were fabricated onto an indium tin oxide (ITO)-glass slide by soft lithography. Following the electrospray deposition (ESD) method, selectively patterned lipid films were formed on the conductive surface (Figure 12).156 With a simple hydration process of the dried lipid, they succeeded in forming giant liposomes on top of the pattern. Although ESD could prepare monodisperse giant vesicles, the area of the sprayed films is too small at once to obtain a high yield.
 |
Figure 12 Electrospray deposition method. (A) Image of a lipid patterned substrate for gentle hydration. (B) Cross-sectional illustration of lipid hydration. An aqueous solution (MilliQ) was simply infused at the wells. ©2011 IEEE. Reprinted with permission, from Osaki T, Kuribayashi-Shigetomi K, Kawano R, Sasaki H, Takeuchi S. Uniformly-sized giant liposome formation with gentle hydration. In: 2011 IEEE 24th International Conference on Micro Electro Mechanical Systems; 2011; IEEE.156. |
One drawback of hydration is the low EE as vesicles cannot be isolated from materials in the external environment during the formation process.157 Moreover, the process requires the deposition of a lipid film on a substrate followed by time-consuming evaporation, which prevents the continuous preparation of vesicles since lipids cannot be replenished during operation. Other microfluidic methods for preparing liposomes, such as micromixers, are more practical when applied to drug delivery.
Electroformation in Chips
The electroformation method for the production of giant liposomes was first described by Angelova and Dimitrov in 1986.30 Similar to hydration, dried lipid films were prepared on the surface of electrodes. An electric field was then applied to the hydrated films immersed in an aqueous solution, resulting in the formation of giant vesicles. However, the electroformation method has a low yield of monodisperse vesicles and lacks an efficient collection process of prepared liposomes.30,154,158
Kuribayashi et al succeeded in integrating the electroformation method within microfluidic chips.159 Two parallel microfluidic channels formed in the silicone sheet were sandwiched between the ITO glass plates to prepare giant vesicles. Liposomes with different lipid compositions prepared by this method could encapsulate different types of materials. Since they were polydisperse, microcontact printing was applied to control the size distribution of vesicles.160–162 The lipid is deposited on an ITO glass using a polydimethylsiloxane (PDMS) stamp of curtain pattern to prepare giant liposomes of narrow size distribution. Also, Diguet et al prepared giant liposomes with controlled size and narrow size distributions by dragging a lipid solution on a microstructured silicon substrate for electroformation.163,164 Recently, Wang et al integrated the preparation, separation, and collection of liposomes onto a PDMS chip. After electroformation, liposomes were driven by fresh buffer and trapped by filter membranes in a tailor-made collection chamber, which increased the liposome collection ratio to 40%.165
In 2013, Bi et al introduced the use of coplanar-interdigitated electrodes for electroformation of liposomes (Figure 13), breaking from the tradition of using two opposite electrodes.166 Smaller interdigitated electrodes generate bigger GUVs in this method, which provides a reliable solution for controlling the size distribution of giant liposomes by electroformation.
 |
Figure 13 Schematic illustration of the GUV electroformation setup by coplanar interdigitated electrode. (A) The interdigitated ITO electrodes (bottom) and a glass coverslip (top) were separated by a PTFE spacer. (B) Side view of the GUV electroformation setup. (C) The actual size of the interdigitated ITO electrodes (top view). Not to scale. Republished with permission of Royal Society of Chemistry from Bi H, Yang B, Wang L, Cao W, Han X. Electroformation of giant unilamellar vesicles using interdigitated ITO electrodes. J Mater Chem A. 2013;1(24):7125–7130. Permission conveyed through the copyright clearance center, Inc.166 |
Apart from those defects discussed in the hydration method, irregular and asymmetric lipid distribution was observed in GUVs formed by electroformation.167 The electric field itself may have adverse effects on liposomes described in Micromixer.98,99 Recent research has mostly focused on exploring novel microfluidics-based electroformation methods for liposomes preparation, and few related applications have been reported,167,168 to some extent implying that applications of electroforming methods are limited due to the shortcomings described above.
Conclusion and Outlook
With the successful development of micro- and nanofabrication,169 the preparation of liposomes has been transposed from the macro- to the micro-scale. Microfluidic technologies can not only precisely control the size and lamellarity of liposomes but also reduce the volume and cost of reagents required simultaneously. Comparison between various microfluidic methods used to prepare liposomes is briefly summarized in Table 2. Different methods have their pros and cons. Thin-film hydration and electroformation are more biocompatible, avoiding the presence of organic solvents. The issue of low EE is insignificant when applied to membrane science. Although the pulse flow jetting and single emulsion templates with higher EE are more difficult to implement, they are near-perfect techniques for synthetic cells. Double emulsion templates, especially the OLA method, are also useful tools to prepare GUVs to explore new drugs and elucidate cellular functions. For the large-scale production of liposomes/LNPs for drug delivery, micromixers may be preferred due to their high throughput.
 |
Table 2 Comparison Between Microfluidic Methods for Liposome Production |
In the case of single emulsion templates, combining dsGUVs with pico injection is one of the few means of directly encapsulating any desired materials into liposomes with the assistance of pico injection technology, which circumvents dilution or contamination of encapsulated materials. Obviously, LNPs are not suitable for larger macromolecules with radii of gyration as large as 1 μm, which may be required for potential applications, such as drug delivery and gene therapy. Moreover, their various strategies for biofunctionalization of GUVs (charged, bioligand-conjugated or PEG-conjugated) widened the horizons in applications of GUVs, such as targeted cellular delivery. Potentially immunogenic cargo could be intracellularly discharged via the lysosomal escape mechanism once the carriers arrive at their destination without initiating an immune response. Work by Spatz group presented an exquisite model for combining liposomes and microfluidic techniques,107,109 motivating further fundamental and applied liposomes research. Coincidentally, several studies have been utilizing other external energies such as acoustic waves and electromagnetics to boost current microfluidic methods by enhancing production efficiency96,97 or simplifying the manufacture of equipment.151 These initiatives have supported the recent efforts to investigate and improve nanotechnology for liposomes formation, but future investigations must critically evaluate whether the properties of liposomes are prone to be disrupted by physical methods like ultrasound and electric field.
No matter which method, bulk or microfluidic, it is necessary to separate and purify liposomes after preparation before further applications. Some microfluidic methods, such as single and double emulsion templates, may require additional post-processing steps (Table 2). For example, the floating oil droplets formed by quick dewetting of the double-emulsion droplets had to be removed by an extra step of pipetting or evaporating.137 OLA also suffered from the presence of 1-octanol droplets, an inevitable by-product of the production process.135 To separate the 1-octanol droplets from the liposomes formed by OLA, a one-step density-based separation technique integrated on the same chip had been developed by the same group.170 A high separation efficiency (~93%) was realized compared to the non-separated samples (~45%). Similarly, Dimov et al introduced a microfluidic system composed of SHM and a tangential flow filtration device for consecutive purification. This on-chip platform can eliminate most non-encapsulated materials and organic solvent (>90%) in less than 4 minutes, offering simultaneous production and purification of liposomes.171 Although purification steps (described in Encapsulation Efficiency) could be conducted off-chip, retaining the liposomes inside the microfluidic chip is of great significance. It reduces purification time, eliminates the possible loss of material during off-chip handling, and, more importantly, enables downstream on-chip experimentation. In fact, endeavors have been made to establish one-step microfluidic platforms for subsequent observation, purification or manipulation of liposomes, such as capture and immobilization,116,172 separations and purification,165 dynamic monitoring173 and even functional evaluation.138,174 These on-chip efforts will certainly broaden the horizons for the research of liposomes/LNPs.
LNPs, an upgraded version of liposomes, have now been utilized as a versatile nanomedicine delivery platform. Although LNPs are currently in the spotlight because of the success of the COVID-19 mRNA vaccines, viral vectors are more frequently used than LNPs since they are more efficient in delivering DNA. RNA only needs to enter the cytoplasm to function, while gene therapy demands the delivery of DNA into the nucleus through the nuclear membrane. Effectively delivering DNA molecules in vitro and in vivo using LNPs requires more in-depth investigations of lipid nanoparticle–DNA formulations.
Most liposomes/LNPs are undoubtedly used in medical science, but academics who usually use these nanoparticles for drug delivery prefer conventional bulk methods, such as ethanol dilution, solvent injection or homogenization to prepare liposomes.19,175,176 They concentrate on designing and optimizing the nanocarrier itself instead of novel techniques for nanocarrier preparation. Since the production techniques cannot resolve the critical clinical problems in liposomal applications, such as delivery efficacy, organ-specific and cell-specific delivery and toxicity, to mention a few, it was not surprising that these bulk methods were chosen due to simplicity, cost-saving and safety. However, researchers that promote the development of preparation techniques pay more attention to the basic design of microfluidic devices like the geometry or the fundamental physics behind the microfluidics-assisted nanoparticle productions, such as the dynamics of the process.177 The blossoming of liposomes/LNPs requires the synergistic development of miscellaneous fields, including medical, physics, chemistry and engineering. In light of this review, we encourage researchers to not only choose the most suitable techniques for the desired research but also bravely explore possible applications of newer techniques. This will likely change in the near future as the popularity of commercial micromixers (eg, the NanoAssemblr™ platform) increased.178,179 Based on the current progress and success, we firmly believe that microfluidics will soon revolutionize the generation of sophisticated artificial cells and lipid-based nanoencapsulation.
Acknowledgments
The authors thank Dr. Zhenxing Wang for his constructive comments during the preparation of this manuscript. This work was supported by the National Key R&D Program of China (2019YFA0110500) and the National Natural Science Foundation of China (No. 8201001114).
Disclosure
The authors have no conflicts of interest to declare that are relevant to the content of this article.
References
1. Bangham AD, Horne RW. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J Mol Biol. 1964;8(5):660–IN610. doi:10.1016/S0022-2836(64)80115-7
2. Bangham AD, Standish MM, Watkins JC. Diffusion of univalent ions across the lamellae of swollen phospholipids. J Mol Biol. 1965;13(1):238–IN227. doi:10.1016/S0022-2836(65)80093-6
3. Nakamura T, Miyabe H, Hyodo M, Sato Y, Hayakawa Y, Harashima H. Liposomes loaded with a STING pathway ligand, cyclic di-GMP, enhance cancer immunotherapy against metastatic melanoma. J Control Release. 2015;216:149–157. doi:10.1016/j.jconrel.2015.08.026
4. Kulkarni JA, Myhre JL, Chen S, et al. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed Nanotechnol Biol Med. 2017;13(4):1377–1387. doi:10.1016/j.nano.2016.12.014
5. Deng H, Song K, Zhao X, et al. Tumor microenvironment activated membrane fusogenic liposome with speedy antibody and doxorubicin delivery for synergistic treatment of metastatic tumors. ACS Appl Mater Interfaces. 2017;9(11):9315–9326. doi:10.1021/acsami.6b14683
6. Lechanteur A, Sanna V, Duchemin A, Evrard B, Mottet D, Piel G. Cationic liposomes carrying siRNA: impact of lipid composition on physicochemical properties, cytotoxicity and endosomal escape. Nanomaterials. 2018;8(5):270. doi:10.3390/nano8050270
7. Elani Y, Law RV, Ces O. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat Commun. 2014;5(1):5305. doi:10.1038/ncomms6305
8. Kaddah S, Khreich N, Kaddah F, Charcosset C, Greige-Gerges H. Cholesterol modulates the liposome membrane fluidity and permeability for a hydrophilic molecule. Food Chem Toxicol. 2018;113:40–48. doi:10.1016/j.fct.2018.01.017
9. Crone NSA, Minnee D, Kros A, Boyle AL. Peptide-mediated liposome fusion: the effect of anchor positioning. Int J Mol Sci. 2018;19(1):211. doi:10.3390/ijms19010211
10. Elani Y, Law RV, Ces O. Protein synthesis in artificial cells: using compartmentalisation for spatial organisation in vesicle bioreactors. Phys Chem Chem Phys. 2015;17(24):15534–15537. doi:10.1039/C4CP05933F
11. Löffler PMG, Ries O, Rabe A, et al. A DNA-programmed liposome fusion cascade. Angewandte Chemie Int Edn. 2017;56(43):13228–13231. doi:10.1002/anie.201703243
12. van Nies P, Westerlaken I, Blanken D, Salas M, Mencía M, Danelon C. Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat Commun. 2018;9(1):1583. doi:10.1038/s41467-018-03926-1
13. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi:10.1056/NEJMoa2034577
14. Khurana A, Allawadhi P, Khurana I, et al. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today. 2021;38:101142. doi:10.1016/j.nantod.2021.101142
15. Butts C, Maksymiuk A, Goss G, et al. Updated survival analysis in patients with stage IIIB or IV non-small-cell lung cancer receiving BLP25 liposome vaccine (L-BLP25): phase IIB randomized, multicenter, open-label trial. J Cancer Res Clin Oncol. 2011;137(9):1337–1342. doi:10.1007/s00432-011-1003-3
16. Yang -Z-Z, Li J-Q, Wang -Z-Z, Dong D-W, Qi X-R. Tumor-targeting dual peptides-modified cationic liposomes for delivery of siRNA and docetaxel to gliomas. Biomaterials. 2014;35(19):5226–5239. doi:10.1016/j.biomaterials.2014.03.017
17. Huang Q, Ji K, Tian S, et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat Commun. 2021;12(1):776. doi:10.1038/s41467-021-21037-2
18. Huang W-C, Zhou S, He X, et al. SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv Mater. 2020;32(50):2005637. doi:10.1002/adma.202005637
19. Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat Nanotechnol. 2020;15(4):313–320. doi:10.1038/s41565-020-0669-6
20. Liu S, Cheng Q, Wei T, et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat Mater. 2021;20(5):701–710. doi:10.1038/s41563-020-00886-0
21. Deng -N-N, Yelleswarapu M, Zheng L, Huck WTS. Microfluidic assembly of monodisperse vesosomes as artificial cell models. J Am Chem Soc. 2017;139(2):587–590. doi:10.1021/jacs.6b10977
22. Deng -N-N, Huck WTS. Microfluidic formation of monodisperse coacervate organelles in liposomes. Angewandte Chemie Int Edn. 2017;56(33):9736–9740. doi:10.1002/anie.201703145
23. Langton MJ, Keymeulen F, Ciaccia M, Williams NH, Hunter CA. Controlled membrane translocation provides a mechanism for signal transduction and amplification. Nat Chem. 2017;9(5):426–430. doi:10.1038/nchem.2678
24. Otake K, Shimomura T, Goto T, et al. Preparation of liposomes using an improved supercritical reverse phase evaporation method. Langmuir. 2006;22(6):2543–2550. doi:10.1021/la051654u
25. Gouda A, Sakr OS, Nasr M, Sammour O. Ethanol injection technique for liposomes formulation: an insight into development, influencing factors, challenges and applications. J Drug Deliv Sci Technol. 2021;61:102174. doi:10.1016/j.jddst.2020.102174
26. Nomura S-M, Tsumoto K, Hamada T, Akiyoshi K, Nakatani Y, Yoshikawa K. Gene expression within cell-sized lipid vesicles. ChemBioChem. 2003;4(11):1172–1175. doi:10.1002/cbic.200300630
27. Kučerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J. 2005;88(4):2626–2637. doi:10.1529/biophysj.104.056606
28. Berger N, Sachse A, Bender J, Schubert R, Brandl M. Filter extrusion of liposomes using different devices: comparison of liposome size, encapsulation efficiency, and process characteristics. Int J Pharm. 2001;223(1):55–68. doi:10.1016/S0378-5173(01)00721-9
29. Jousma H, Talsma H, Spies F, Joosten JGH, Junginger HE, Crommelin DJA. Characterization of liposomes. The influence of extrusion of multilamellar vesicles through polycarbonate membranes on particle size, particle size distribution and number of bilayers. Int J Pharm. 1987;35(3):263–274. doi:10.1016/0378-5173(87)90139-6
30. Angelova MI, Dimitrov DS. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303–311.
31. Estes DJ, Mayer M. Electroformation of giant liposomes from spin-coated films of lipids. Colloids Surf B Biointerfaces. 2005;42(2):115–123. doi:10.1016/j.colsurfb.2005.01.016
32. Li C, Deng Y. A novel method for the preparation of liposomes: freeze drying of monophase solutions. J Pharm Sci. 2004;93(6):1403–1414. doi:10.1002/jps.20055
33. Cui J, Li C, Deng Y, Wang Y, Wang W. Freeze-drying of liposomes using tertiary butyl alcohol/water cosolvent systems. Int J Pharm. 2006;312(1):131–136. doi:10.1016/j.ijpharm.2006.01.004
34. Pautot S, Frisken BJ, Weitz DA. Engineering asymmetric vesicles. Proc Natl Acad Sci. 2003;100(19):10718. doi:10.1073/pnas.1931005100
35. Liau JJ, Hook S, Prestidge CA, Barnes TJ. A lipid based multi-compartmental system: liposomes-in-double emulsion for oral vaccine delivery. Eur J Pharma Biopharm. 2015;97:15–21. doi:10.1016/j.ejpb.2015.09.018
36. Nagayasu A, Uchiyama K, Kiwada H. The size of liposomes: a factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv Drug Deliv Rev. 1999;40(1–2):75–87. doi:10.1016/S0169-409X(99)00041-1
37. Cabral H, Matsumoto Y, Mizuno K, et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat Nanotechnol. 2011;6(12):815–823. doi:10.1038/nnano.2011.166
38. Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65(1):36–48.
39. van Swaay D, deMello A. Microfluidic methods for forming liposomes. Lab Chip. 2013;13(5):752–767. doi:10.1039/c2lc41121k
40. Carugo D, Bottaro E, Owen J, Stride E, Nastruzzi C. Liposome production by microfluidics: potential and limiting factors. Sci Rep. 2016;6(1):25876. doi:10.1038/srep25876
41. Has C, Sunthar P. A comprehensive review on recent preparation techniques of liposomes. J Liposome Res. 2020;30(4):336–365. doi:10.1080/08982104.2019.1668010
42. Akbarzadeh A, Rezaei-Sadabady R, Davaran S, et al. Liposome: classification, preparation, and applications. Nanoscale Res Lett. 2013;8(1):102. doi:10.1186/1556-276X-8-102
43. Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi:10.1007/s11095-010-0073-2
44. Markones M, Drechsler C, Kaiser M, Kalie L, Heerklotz H, Fiedler S. Engineering asymmetric lipid vesicles: accurate and convenient control of the outer leaflet lipid composition. Langmuir. 2018;34(5):1999–2005. doi:10.1021/acs.langmuir.7b03189
45. Villasmil-Sánchez S, Rabasco AM, González-Rodríguez ML. Thermal and 31P-NMR studies to elucidate sumatriptan succinate entrapment behavior in phosphatidylcholine/cholesterol liposomes. Comparative 31P-NMR analysis on negatively and positively-charged liposomes. Colloids Surf B Biointerfaces. 2013;105:14–23. doi:10.1016/j.colsurfb.2012.12.019
46. Ghazal A, Gontsarik M, Kutter JP, et al. Microfluidic platform for the continuous production and characterization of multilamellar vesicles: a synchrotron small-angle X-ray scattering (SAXS) study. J Phys Chem Lett. 2017;8(1):73–79. doi:10.1021/acs.jpclett.6b02468
47. Hoppe SM, Sasaki DY, Kinghorn AN, Hattar K. In-situ transmission electron microscopy of liposomes in an aqueous environment. Langmuir. 2013;29(32):9958–9961. doi:10.1021/la401288g
48. Cohen SM. Extended pain relief trial utilizing infiltration of Exparel(®), a long-acting multivesicular liposome formulation of bupivacaine: a phase IV health economic trial in adult patients undergoing open colectomy. J Pain Res. 2012;5:567–572. doi:10.2147/JPR.S38621
49. Kao H-W, Lin -Y-Y, Gwathney WJ, Hong K. Formulation and evaluation of multilamellar vesicles ropivacaine in pain management. Int J Nanomedicine. 2019;14:7891–7901. doi:10.2147/IJN.S215952
50. Liu D, Mori A, Huang L. Role of liposome size and RES blockade in controlling biodistribution and tumor uptake of GM1-containing liposomes. Biochim Biophys Acta. 1992;1104(1):95–101. doi:10.1016/0005-2736(92)90136-A
51. Jensen GM, Hodgson DF. Opportunities and challenges in commercial pharmaceutical liposome applications. Adv Drug Del Rev. 2020;154–155:2–12. doi:10.1016/j.addr.2020.07.016
52. Robinson T. Microfluidic handling and analysis of giant vesicles for use as artificial cells: a review. Adv Biosyst. 2019;3(6):1800318. doi:10.1002/adbi.201800318
53. Briuglia M-L, Rotella C, McFarlane A, Lamprou DA. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Deliv Transl Res. 2015;5(3):231–242. doi:10.1007/s13346-015-0220-8
54. Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163(3):267–276. doi:10.1016/j.jconrel.2012.09.009
55. Moon JJ, Suh H, Bershteyn A, et al. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat Mater. 2011;10(3):243–251. doi:10.1038/nmat2960
56. Zhang Y, Mintzer E, Uhrich KE. Synthesis and characterization of PEGylated bolaamphiphiles with enhanced retention in liposomes. J Colloid Interface Sci. 2016;482:19–26. doi:10.1016/j.jcis.2016.07.013
57. Caddeo C, Pucci L, Gabriele M, et al. Stability, biocompatibility and antioxidant activity of PEG-modified liposomes containing resveratrol. Int J Pharm. 2018;538(1–2):40–47. doi:10.1016/j.ijpharm.2017.12.047
58. Lombardo D, Calandra P, Barreca D, Magazù S, Kiselev MA. Soft interaction in liposome nanocarriers for therapeutic drug delivery. Nanomaterials. 2016;6(7):125. doi:10.3390/nano6070125
59. Narenji M, Talaee MR, Moghimi HR. Effect of charge on separation of liposomes upon stagnation. Iran J Pharm Res. 2017;16(2):423–431.
60. Clogston JD, Patri AK. Zeta potential measurement. In: Characterization of Nanoparticles Intended for Drug Delivery. Humana Press; 2011:63–70.
61. Gonzalez Gomez A, Syed S, Marshall K, Hosseinidoust Z. Liposomal nanovesicles for efficient encapsulation of staphylococcal antibiotics. ACS Omega. 2019;4(6):10866–10876. doi:10.1021/acsomega.9b00825
62. Ruysschaert T, Marque A, Duteyrat J-L, Lesieur S, Winterhalter M, Fournier D. Liposome retention in size exclusion chromatography. BMC Biotechnol. 2005;5:11. doi:10.1186/1472-6750-5-11
63. Ran C, Chen D, Xu M, Du C, Li Q, Jiang Y. A study on characteristic of different sample pretreatment methods to evaluate the entrapment efficiency of liposomes. J Chromatogr B. 2016;1028:56–62. doi:10.1016/j.jchromb.2016.06.008
64. Zhang XM, Patel AB, de Graaf RA, Behar KL. Determination of liposomal encapsulation efficiency using proton NMR spectroscopy. Chem Phys Lipids. 2004;127(1):113–120. doi:10.1016/j.chemphyslip.2003.09.013
65. Lohse B, Bolinger P-Y, Stamou D. Encapsulation efficiency measured on single small unilamellar vesicles. J Am Chem Soc. 2008;130(44):14372–14373. doi:10.1021/ja805030w
66. Sun B, Chiu DT. Determination of the encapsulation efficiency of individual vesicles using single-vesicle photolysis and confocal single-molecule detection. Anal Chem. 2005;77(9):2770–2776. doi:10.1021/ac048439n
67. Yamamoto E, Miyazaki S, Aoyama C, Kato M. A simple and rapid measurement method of encapsulation efficiency of doxorubicin loaded liposomes by direct injection of the liposomal suspension to liquid chromatography. Int J Pharm. 2018;536(1):21–28. doi:10.1016/j.ijpharm.2017.11.035
68. Jahn A, Vreeland WN, Gaitan M, Locascio LE. Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J Am Chem Soc. 2004;126(9):2674–2675. doi:10.1021/ja0318030
69. Jahn A, Vreeland WN, DeVoe DL, Locascio LE, Gaitan M. Microfluidic directed formation of liposomes of controlled size. Langmuir. 2007;23(11):6289–6293. doi:10.1021/la070051a
70. Jahn A, Stavis SM, Hong JS, Vreeland WN, DeVoe DL, Gaitan M. Microfluidic mixing and the formation of nanoscale lipid vesicles. ACS Nano. 2010;4(4):2077–2087. doi:10.1021/nn901676x
71. Tien Sing Young RV, Tabrizian M. Rapid, one-step fabrication and loading of nanoscale 1,2-distearoyl-sn-glycero-3-phosphocholine liposomes in a simple, double flow-focusing microfluidic device. Biomicrofluidics. 2015;9(4):046501. doi:10.1063/1.4926398
72. Mijajlovic M, Wright D, Zivkovic V, Bi JX, Biggs MJ. Microfluidic hydrodynamic focusing based synthesis of POPC liposomes for model biological systems. Colloids Surf B Biointerfaces. 2013;104:276–281. doi:10.1016/j.colsurfb.2012.12.020
73. Zizzari A, Bianco M, Carbone L, et al. Continuous-flow production of injectable liposomes via a microfluidic approach. Materials. 2017;10(12):1411. doi:10.3390/ma10121411
74. Jahn A, Lucas F, Wepf RA, Dittrich PS. Freezing continuous-flow self-assembly in a microfluidic device: toward imaging of liposome formation. Langmuir. 2013;29(5):1717–1723. doi:10.1021/la303675g
75. Balbino TA, Aoki NT, Gasperini AAM, et al. Continuous flow production of cationic liposomes at high lipid concentration in microfluidic devices for gene delivery applications. Chem Eng J. 2013;226:423–433. doi:10.1016/j.cej.2013.04.053
76. Aghaei H, Solaimany Nazar AR. Continuous production of the nanoscale liposome in a double flow-focusing microfluidic device. Ind Eng Chem Res. 2019;58(51):23032–23045. doi:10.1021/acs.iecr.9b04079
77. Hood RR, DeVoe DL, Atencia J, Vreeland WN, Omiatek DM. A facile route to the synthesis of monodisperse nanoscale liposomes using 3D microfluidic hydrodynamic focusing in a concentric capillary array. Lab Chip. 2014;14(14):2403–2409. doi:10.1039/C4LC00334A
78. Hood RR, DeVoe DL. High-throughput continuous flow production of nanoscale liposomes by microfluidic vertical flow focusing. Small. 2015;11(43):5790–5799. doi:10.1002/smll.201501345
79. Michelon M, Oliveira DRB, de Figueiredo Furtado G, Gaziola de la Torre L, Cunha RL. High-throughput continuous production of liposomes using hydrodynamic flow-focusing microfluidic devices. Colloids Surf B Biointerfaces. 2017;156:349–357. doi:10.1016/j.colsurfb.2017.05.033
80. Chen Z, Han JY, Shumate L, Fedak R, DeVoe DL. High throughput nanoliposome formation using 3D printed microfluidic flow focusing chips. Adv Mater Technol. 2019;4(6):1800511. doi:10.1002/admt.201800511
81. Balbino TA, Azzoni AR, de la Torre LG. Microfluidic devices for continuous production of pDNA/cationic liposome complexes for gene delivery and vaccine therapy. Colloids Surf B Biointerfaces. 2013;111:203–210. doi:10.1016/j.colsurfb.2013.04.003
82. Ran R, Middelberg APJ, Zhao C-X. Microfluidic synthesis of multifunctional liposomes for tumour targeting. Colloids Surf B Biointerfaces. 2016;148:402–410. doi:10.1016/j.colsurfb.2016.09.016
83. Aghaei H, Nazar ARS, Varshosaz J. Double flow focusing microfluidic-assisted based preparation of Methotrexate–Loaded liposomal nanoparticles: encapsulation efficacy, drug release and stability. Colloids Surf Physicochem Eng Aspects. 2021;614:126166. doi:10.1016/j.colsurfa.2021.126166
84. Hood RR, Vreeland WN, DeVoe DL. Microfluidic remote loading for rapid single-step liposomal drug preparation. Lab Chip. 2014;14(17):3359–3367. doi:10.1039/C4LC00390J
85. Sur S, Fries AC, Kinzler KW, Zhou S, Vogelstein B. Remote loading of preencapsulated drugs into stealth liposomes. Proc Natl Acad Sci. 2014;111(6):2283. doi:10.1073/pnas.1324135111
86. Stroock AD, Dertinger SKW, Ajdari A, Mezić I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science. 2002;295(5555):647. doi:10.1126/science.1066238
87. Zhigaltsev IV, Belliveau N, Hafez I, et al. Bottom-up design and synthesis of limit size lipid nanoparticle systems with aqueous and triglyceride cores using millisecond microfluidic mixing. Langmuir. 2012;28(7):3633–3640. doi:10.1021/la204833h
88. Kastner E, Kaur R, Lowry D, Moghaddam B, Wilkinson A, Perrie Y. High-throughput manufacturing of size-tuned liposomes by a new microfluidics method using enhanced statistical tools for characterization. Int J Pharm. 2014;477(1):361–368. doi:10.1016/j.ijpharm.2014.10.030
89. Maeki M, Saito T, Sato Y, et al. A strategy for synthesis of lipid nanoparticles using microfluidic devices with a mixer structure. RSC Adv. 2015;5(57):46181–46185. doi:10.1039/C5RA04690D
90. Lou G, Anderluzzi G, Woods S, Roberts CW, Perrie Y. A novel microfluidic-based approach to formulate size-tuneable large unilamellar cationic liposomes: formulation, cellular uptake and biodistribution investigations. Eur J Pharma Biopharm. 2019;143:51–60. doi:10.1016/j.ejpb.2019.08.013
91. Maeki M, Fujishima Y, Sato Y, et al. Understanding the formation mechanism of lipid nanoparticles in microfluidic devices with chaotic micromixers. PLoS One. 2017;12(11):e0187962. doi:10.1371/journal.pone.0187962
92. Joshi S, Hussain MT, Roces CB, et al. Microfluidics based manufacture of liposomes simultaneously entrapping hydrophilic and lipophilic drugs. Int J Pharm. 2016;514(1):160–168. doi:10.1016/j.ijpharm.2016.09.027
93. Tenchov R, Bird R, Curtze AE, Zhou Q. Lipid nanoparticles—from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021. doi:10.1021/acsnano.1c04996
94. Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20(2):101–124. doi:10.1038/s41573-020-0090-8
95. Kimura N, Maeki M, Sato Y, et al. Development of the iLiNP device: fine tuning the lipid nanoparticle size within 10 nm for drug delivery. ACS Omega. https://pubs.acs.org/doi/10.1021/acsomega.8b00341. 2018;3(5):5044–5051.
96. Rasouli MR, Tabrizian M. An ultra-rapid acoustic micromixer for synthesis of organic nanoparticles. Lab Chip. 2019;19(19):3316–3325. doi:10.1039/C9LC00637K
97. Modarres P, Tabrizian M. Electrohydrodynamic-driven micromixing for the synthesis of highly monodisperse nanoscale liposomes. ACS Appl Nano Mater. 2020;3(5):4000–4013. doi:10.1021/acsanm.9b02407
98. Breton M, Amirkavei M, Mir LM. Optimization of the electroformation of giant unilamellar vesicles (GUVs) with unsaturated phospholipids. J Membr Biol. 2015;248(5):827–835. doi:10.1007/s00232-015-9828-3
99. Runas KA, Malmstadt N. Low levels of lipid oxidation radically increase the passive permeability of lipid bilayers. Soft Matter. 2015;11(3):499–505. doi:10.1039/C4SM01478B
100. Kastner E, Verma V, Lowry D, Perrie Y. Microfluidic-controlled manufacture of liposomes for the solubilisation of a poorly water soluble drug. Int J Pharm. 2015;485(1):122–130. doi:10.1016/j.ijpharm.2015.02.063
101. Chen D, Love KT, Chen Y, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134(16):6948–6951. doi:10.1021/ja301621z
102. Belliveau NM, Huft J, Lin PJ, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1(8):e37. doi:10.1038/mtna.2012.28
103. Leung AKK, Hafez IM, Baoukina S, et al. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J Phys Chem C. 2012;116(34):18440–18450. doi:10.1021/jp303267y
104. Shepherd SJ, Warzecha CC, Yadavali S, et al. Scalable mRNA and siRNA lipid nanoparticle production using a parallelized microfluidic device. Nano Lett. 2021;21(13):5671–5680. doi:10.1021/acs.nanolett.1c01353
105. Rosenblum D, Gutkin A, Kedmi R, et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020;6(47):eabc9450. doi:10.1126/sciadv.abc9450
106. Riley RS, Kashyap MV, Billingsley MM, et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci Adv. 2021;7(3). doi:10.1126/sciadv.aba1028.
107. Weiss M, Frohnmayer JP, Benk LT, et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat Mater. 2018;17(1):89–96. doi:10.1038/nmat5005
108. Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA. High-throughput injection with microfluidics using picoinjectors. Proc Natl Acad Sci. 2010;107(45):19163. doi:10.1073/pnas.1006888107
109. Staufer O, Antona S, Zhang D, et al. Microfluidic production and characterization of biofunctionalized giant unilamellar vesicles for targeted intracellular cargo delivery. Biomaterials. 2021;264:120203. doi:10.1016/j.biomaterials.2020.120203
110. Sugiura S, Kuroiwa T, Kagota T, et al. Novel method for obtaining homogeneous giant vesicles from a monodisperse water-in-oil emulsion prepared with a microfluidic device. Langmuir. 2008;24(9):4581–4588. doi:10.1021/la703509r
111. Kuroiwa T, Fujita R, Kobayashi I, et al. Efficient preparation of giant vesicles as biomimetic compartment systems with high entrapment yields for biomacromolecules. Chem Biodivers. 2012;9(11):2453–2472. doi:10.1002/cbdv.201200274
112. Pautot S, Frisken BJ, Weitz DA. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19(7):2870–2879. doi:10.1021/la026100v
113. Tan Y-C, Hettiarachchi K, Siu M, Pan Y-R, Lee AP. Controlled microfluidic encapsulation of cells, proteins, and microbeads in lipid vesicles. J Am Chem Soc. 2006;128(17):5656–5658. doi:10.1021/ja056641h
114. Hu PC, Li S, Malmstadt N. Microfluidic fabrication of asymmetric giant lipid vesicles. ACS Appl Mater Interfaces. 2011;3(5):1434–1440. doi:10.1021/am101191d
115. Nishimura K, Suzuki H, Toyota T, Yomo T. Size control of giant unilamellar vesicles prepared from inverted emulsion droplets. J Colloid Interface Sci. 2012;376(1):119–125. doi:10.1016/j.jcis.2012.02.029
116. Matosevic S, Paegel BM. Stepwise synthesis of giant unilamellar vesicles on a microfluidic assembly line. J Am Chem Soc. 2011;133(9):2798–2800. doi:10.1021/ja109137s
117. Karamdad K, Law RV, Seddon JM, Brooks NJ, Ces O. Preparation and mechanical characterisation of giant unilamellar vesicles by a microfluidic method. Lab Chip. 2015;15(2):557–562. doi:10.1039/C4LC01277A
118. Karamdad K, Law RV, Seddon JM, Brooks NJ, Ces O. Studying the effects of asymmetry on the bending rigidity of lipid membranes formed by microfluidics. Chem Commun. 2016;52(30):5277–5280. doi:10.1039/C5CC10307J
119. Matosevic S, Paegel BM. Layer-by-layer cell membrane assembly. Nat Chem. 2013;5(11):958–963. doi:10.1038/nchem.1765
120. Morita M, Onoe H, Yanagisawa M, et al. Droplet-shooting and size-filtration (DSSF) method for synthesis of cell-sized liposomes with controlled lipid compositions. ChemBioChem. 2015;16(14):2029–2035. doi:10.1002/cbic.201500354
121. Morita M, Katoh K, Noda N. Direct observation of bacterial growth in giant unilamellar vesicles: a novel tool for bacterial cultures. ChemistryOpen. 2018;7(11):845–849. doi:10.1002/open.201800126
122. Adir O, Sharf-Pauker N, Chen G, et al. Preparing protein producing synthetic cells using cell free bacterial extracts, liposomes and emulsion transfer. JoVE. 2020;158:e60829.
123. Kubatta EA, Rehage H. Characterization of giant vesicles formed by phase transfer processes. Colloid Polym Sci. 2009;287(9):1117–1122. doi:10.1007/s00396-009-2083-3
124. Shum HC, Lee D, Yoon I, Kodger T, Weitz DA. Double emulsion templated monodisperse phospholipid vesicles. Langmuir. 2008;24(15):7651–7653. doi:10.1021/la801833a
125. Kim S-H, Kim JW, Cho J-C, Weitz DA. Double-emulsion drops with ultra-thin shells for capsule templates. Lab Chip. 2011;11(18):3162–3166. doi:10.1039/C1LC20434C
126. Arriaga LR, Datta SS, Kim SH, et al. Ultrathin shell double emulsion templated giant unilamellar lipid vesicles with controlled microdomain formation. Small. 2014;10(5):950–956. doi:10.1002/smll.201301904
127. Caschera F, Lee JW, Ho KKY, Liu AP, Jewett MC. Cell-free compartmentalized protein synthesis inside double emulsion templated liposomes with in vitro synthesized and assembled ribosomes. Chem Commun. 2016;52(31):5467–5469. doi:10.1039/C6CC00223D
128. Costa C, Liu Z, Simões SI, et al. One-step microfluidics production of enzyme-loaded liposomes for the treatment of inflammatory diseases. Colloids Surf B Biointerfaces. 2021;199:111556. doi:10.1016/j.colsurfb.2020.111556
129. Teh S-Y, Khnouf R, Fan H, Lee AP. Stable, biocompatible lipid vesicle generation by solvent extraction-based droplet microfluidics. Biomicrofluidics. 2011;5(4):44113–4411312. doi:10.1063/1.3665221
130. Petit J, Polenz I, Baret J-C, Herminghaus S, Bäumchen O. Vesicles-on-a-chip: a universal microfluidic platform for the assembly of liposomes and polymersomes. Eur Phys J E. 2016;39(6):59. doi:10.1140/epje/i2016-16059-8
131. Lu L, Doak WJ, Schertzer JW, Chiarot PR. Membrane mechanical properties of synthetic asymmetric phospholipid vesicles. Soft Matter. 2016;12(36):7521–7528. doi:10.1039/C6SM01349J
132. Arriaga LR, Huang Y, Kim S-H, et al. Single-step assembly of asymmetric vesicles. Lab Chip. 2019;19(5):749–756. doi:10.1039/C8LC00882E
133. Lu L, Schertzer JW, Chiarot PR. Continuous microfluidic fabrication of synthetic asymmetric vesicles. Lab Chip. 2015;15(17):3591–3599. doi:10.1039/C5LC00520E
134. Lee JN, Park C, Whitesides GM. Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Anal Chem. 2003;75(23):6544–6554. doi:10.1021/ac0346712
135. Deshpande S, Caspi Y, Meijering AE, Dekker C. Octanol-assisted liposome assembly on chip. Nat Commun. 2016;7:10447. doi:10.1038/ncomms10447
136. Deshpande S, Dekker C. On-chip microfluidic production of cell-sized liposomes. Nat Protoc. 2018;13(5):856–874. doi:10.1038/nprot.2017.160
137. Deng -N-N, Yelleswarapu M, Huck WTS. Monodisperse uni- and multicompartment liposomes. J Am Chem Soc. 2016;138(24):7584–7591. doi:10.1021/jacs.6b02107
138. Schaich M, Cama J, Al Nahas K, et al. An integrated microfluidic platform for quantifying drug permeation across biomimetic vesicle membranes. Mol Pharm. 2019;16(6):2494–2501. doi:10.1021/acs.molpharmaceut.9b00086
139. Vaezi Z, Sedghi M, Ghorbani M, Shojaeilangari S, Allahverdi A, Naderi-Manesh H. Investigation of the programmed cell death by encapsulated cytoskeleton drug liposomes using a microfluidic platform. Microfluid Nanofluid. 2020;24(7):48. doi:10.1007/s10404-020-02353-3
140. Niederholtmeyer H, Chaggan C, Devaraj NK. Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat Commun. 2018;9(1):5027. doi:10.1038/s41467-018-07473-7
141. Deshpande S, Brandenburg F, Lau A, et al. Spatiotemporal control of coacervate formation within liposomes. Nat Commun. 2019;10(1):1800. doi:10.1038/s41467-019-09855-x
142. Funakoshi K, Suzuki H, Takeuchi S. Formation of giant lipid vesicle like compartments from a planar lipid membrane by a pulsed jet flow. J Am Chem Soc. 2007;129(42):12608–12609. doi:10.1021/ja074029f
143. Funakoshi K, Suzuki H, Takeuchi S. Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal Chem. 2006;78(24):8169–8174. doi:10.1021/ac0613479
144. Kamiya K, Kawano R, Osaki T, Akiyoshi K, Takeuchi S. Cell-sized asymmetric lipid vesicles facilitate the investigation of asymmetric membranes. Nat Chem. 2016;8(9):881–889. doi:10.1038/nchem.2537
145. Kamiya K, Osaki T, Takeuchi S. Formation of vesicles-in-a-vesicle with asymmetric lipid components using a pulsed-jet flow method. RSC Adv. 2019;9(52):30071–30075. doi:10.1039/C9RA04622D
146. Kamiya K, Osaki T, Takeuchi S. Formation of nano-sized lipid vesicles with asymmetric lipid components using a pulsed-jet flow method. Sensors Actuators B: Chem. 2021;327:128917. doi:10.1016/j.snb.2020.128917
147. Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc Natl Acad Sci. 2008;105(12):4697. doi:10.1073/pnas.0710875105
148. Stachowiak JC, Richmond DL, Li TH, Brochard-Wyart F, Fletcher DA. Inkjet formation of unilamellar lipid vesicles for cell-like encapsulation. Lab Chip. 2009;9(14):2003–2009. doi:10.1039/b904984c
149. Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N, Fletcher DA. Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proc Natl Acad Sci. 2011;108(23):9431. doi:10.1073/pnas.1016410108
150. Gotanda M, Kamiya K, Osaki T, Miki N, Takeuchi S. Automatic generation system of cell-sized liposomes. Sensors Actuators B: Chem. 2019;292:57–63. doi:10.1016/j.snb.2019.04.096
151. Armstrong M, Vahey MD, Hunt TP, Fletcher DA. Forming and loading giant unilamellar vesicles with acoustic jetting. Biomicrofluidics. 2020;14(6):064105. doi:10.1063/5.0021742
152. Kirchner SR, Ohlinger A, Pfeiffer T, et al. Membrane composition of jetted lipid vesicles: a Raman spectroscopy study. J Biophotonics. 2012;5(1):40–46. doi:10.1002/jbio.201100058
153. Reeves JP, Dowben RM. Formation and properties of thin-walled phospholipid vesicles. J Cell Physiol. 1969;73(1):49–60. doi:10.1002/jcp.1040730108
154. Rodriguez N, Pincet F, Cribier S. Giant vesicles formed by gentle hydration and electroformation: a comparison by fluorescence microscopy. Colloids Surf B Biointerfaces. 2005;42(2):125–130. doi:10.1016/j.colsurfb.2005.01.010
155. Zhang H. Thin-film hydration followed by extrusion method for liposome preparation. In: Liposomes. New York, NY: Humana Press; 2017:17–22.
156. Osaki T, Kuribayashi-Shigetomi K, Kawano R, Sasaki H, Takeuchi S. Uniformly-sized giant liposome formation with gentle hydration. In:
157. O’Neil CP, Suzuki T, Demurtas D, Finka A, Hubbell JA. A novel method for the encapsulation of biomolecules into polymersomes via direct hydration. Langmuir. 2009;25(16):9025–9029. doi:10.1021/la900779t
158. Herold C, Chwastek G, Schwille P, Petrov EP. Efficient electroformation of supergiant unilamellar vesicles containing cationic lipids on ITO-coated electrodes. Langmuir. 2012;28(13):5518–5521. doi:10.1021/la3005807
159. Kuribayashi K, Tresset G, Coquet P, Fujita H, Takeuchi S. Electroformation of giant liposomes in microfluidic channels. Meas Sci Technol. 2006;17(12):3121–3126. doi:10.1088/0957-0233/17/12/S01
160. Taylor P, Xu C, Fletcher PDI, Paunov VN. A novel technique for preparation of monodisperse giant liposomes. Chem Commun. 2003;(14):1732–1733. doi:10.1039/b304059c
161. Taylor P, Xu C, Fletcher PDI, Paunov VN. Fabrication of 2D arrays of giant liposomes on solid substrates by microcontact printing. Phys Chem Chem Phys. 2003;5(21):4918–4922. doi:10.1039/b308082j
162. Nafday OA, Lowry TW, Lenhert S. Multifunctional lipid multilayer stamping. Small. 2012;8(7):1021–1028. doi:10.1002/smll.201102096
163. Le Berre M, Yamada A, Reck L, Chen Y, Baigl D. Electroformation of giant phospholipid vesicles on a silicon substrate: advantages of controllable surface properties. Langmuir. 2008;24(6):2643–2649. doi:10.1021/la703391q
164. Diguet A, Le Berre M, Chen Y, Baigl D. Preparation of phospholipid multilayer patterns of controlled size and thickness by capillary assembly on a microstructured substrate. Small. 2009;5(14):1661–1666. doi:10.1002/smll.200900368
165. Wang Z, Wu C, Fan T, et al. Electroformation and collection of giant liposomes on an integrated microchip. Chin Chem Lett. 2019;30(2):353–358. doi:10.1016/j.cclet.2018.12.001
166. Bi H, Yang B, Wang L, Cao W, Han X. Electroformation of giant unilamellar vesicles using interdigitated ITO electrodes. J Mater Chem A. 2013;1(24):7125–7130. doi:10.1039/c3ta10323d
167. Steinkühler J, De Tillieux P, Knorr RL, Lipowsky R, Dimova R. Charged giant unilamellar vesicles prepared by electroformation exhibit nanotubes and transbilayer lipid asymmetry. Sci Rep. 2018;8(1):11838. doi:10.1038/s41598-018-30286-z
168. Witkowska A, Jablonski L, Jahn R. A convenient protocol for generating giant unilamellar vesicles containing SNARE proteins using electroformation. Sci Rep. 2018;8(1):9422. doi:10.1038/s41598-018-27456-4
169. Cheng L, Wang Y, Sun G, et al. Hydration-enhanced lubricating electrospun nanofibrous membranes prevent tissue adhesion. Research. 2020;2020:4907185. doi:10.34133/2020/4907185
170. Deshpande S, Birnie A, Dekker C. On-chip density-based purification of liposomes. Biomicrofluidics. 2017;11(3):034106. doi:10.1063/1.4983174
171. Dimov N, Kastner E, Hussain M, Perrie Y, Szita N. Formation and purification of tailored liposomes for drug delivery using a module-based micro continuous-flow system. Sci Rep. 2017;7(1):12045. doi:10.1038/s41598-017-11533-1
172. Haller B, Göpfrich K, Schröter M, Janiesch J-W, Platzman I, Spatz JP. Charge-controlled microfluidic formation of lipid-based single- and multicompartment systems. Lab Chip. 2018;18(17):2665–2674. doi:10.1039/C8LC00582F
173. Birnbaumer G, Küpcü S, Jungreuthmayer C, et al. Rapid liposome quality assessment using a lab-on-a-chip. Lab Chip. 2011;11(16):2753–2762. doi:10.1039/c0lc00589d
174. Ran R, Wang H-F, Hou F, et al. A microfluidic tumor-on-a-chip for assessing multifunctional liposomes’ tumor targeting and anticancer efficacy. Adv Healthcare Mater. 2019;8(8):1900015. doi:10.1002/adhm.201900015
175. Muthu MS, Kulkarni SA, Raju A, Feng SS. Theranostic liposomes of TPGS coating for targeted co-delivery of docetaxel and quantum dots. Biomaterials. 2012;33(12):3494–3501. doi:10.1016/j.biomaterials.2012.01.036
176. Keck CM, Specht D, Brüßler J. Influence of lipid matrix composition on biopharmaceutical properties of lipid nanoparticles. J Control Release. 2021;338:149–163. doi:10.1016/j.jconrel.2021.08.016
177. Liu Z, Fontana F, Python A, Hirvonen JT, Santos HA. Microfluidics: microfluidics for production of particles: mechanism, methodology, and applications (Small 9/2020). Small. 2020;16(9):2070048. doi:10.1002/smll.202070048
178. Qiu M, Glass Z, Chen J, et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc Natl Acad Sci U S A. 2021;118(10):e2020401118. doi:10.1073/pnas.2020401118
179. Elia U, Ramishetti S, Rosenfeld R, et al. Design of SARS-CoV-2 hFc-conjugated receptor-binding domain mRNA vaccine delivered via lipid nanoparticles. ACS Nano. 2021;15(6):9627–9637. doi:10.1021/acsnano.0c10180
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

