Back to Journals » OncoTargets and Therapy » Volume 11
lincROR influences the stemness and crizotinib resistance in EML–ALK+ non-small-cell lung cancer cells
Authors Yang Y, Huang J, Xie N, Huang H , Xu S, Cai J, Qi S
Received 11 February 2018
Accepted for publication 19 April 2018
Published 22 June 2018 Volume 2018:11 Pages 3649—3657
DOI https://doi.org/10.2147/OTT.S165290
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Yonghua Yang,1,* Jingyu Huang,2,* Nianlin Xie,3,* Hu Huang,4,* Shaogan Xu,5 Jun Cai,1 Shuai Qi6
1Department of Oncology, First Affiliated Hospital of Yangtze University, Jingzhou 434000, Hubei Province, China; 2Department of Thoracic Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, Hubei Province, China; 3Department of Thoracic Surgery, Tangdu Hospital, Fourth Military Medical University, Xi’an 710038, Shaanxi Province, China; 4Department of Oncology, 5Department of Thoracic Surgery, 6Department of Pharmacy, The 161th Hospital of PLA, Wuhan 430010, Hubei Province, China
*These authors contributed equally to this work
Introduction: Echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4–ALK) is identified as an important pathogenic factor in patients with non-small-cell lung cancer (NSCLC) and could induce a stem-like phenotype in NSCLC cells. Crizotinib is commonly used for EML4–ALK+ NSCLC treatment, but its acquired resistance results in tumor recurrence. Long intergenic noncoding RNA, regulator of reprogramming (lincROR) is related to the acquisition and maintenance of self-renewal and stemness features of cancer stem cells. It has been documented that lincROR is implicated in chemoresistance. However, the correlations of lincROR and EML4–ALK in stem cell-like properties and of lincROR and crizotinib resistance in NSCLC cells are yet to be elucidated.
Patients and methods: In the present study, we investigated the expression profile of lincROR in EML–ALK NSCLC tissues, and the potential role of lincROR in prognosis was then analyzed. Subsequently, its association with stem cell-like properties of EML–ALK+ NSCLC cells was determined. Furthermore, the correlation of lincROR with crizotinib and the effects of lincROR and crizotinib on cell viability of EML4–ALK+ NSCLC cells were all explored.
Results: The results showed that lincROR expression was upregulated in EML4–ALK+ NSCLC tissues relative to EML4–ALK- NSCLC tissues. Low-expressed lincROR was related to a favorable prognosis of patients with EML–ALK NSCLC. lincROR overexpression could enhance the stemness features of EML–ALK+ NSCLC cells which were repressed by ALK knockdown.
Conclusion: We found that lincROR expression was significantly inhibited because of the increased concentration of crizotinib in EML4–ALK+ NSCLC cells. Furthermore, lincROR overexpression increased cell viability of EML4–ALK+ NSCLC cells, which was impaired by crizotinib. Conjointly, these results suggested the important role of lincROR in EML–ALK+ NSCLC. lincROR may serve as a potential therapeutic target to overcome chemotherapy resistance in EML–ALK+ NSCLC.
Keywords: EML–ALK, lincROR, stemness, crizotinib, NSCLC
Introduction
Lung cancer remains the most prevalent malignant tumor in the respiratory system with the highest morbidity and mortality.1,2 Non-small-cell lung cancer (NSCLC) serves as a subtype of lung cancer and accounts for ~80%–85% of all lung cancers.3 Because the great majority of NSCLC patients are diagnosed in the middle or terminal stage, clinical drug treatments have become the focus of NSCLC.4 Echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4–ALK) is a transforming gene and a driver mutation in NSCLC, which has been identified to be closely associated with cancerogenesis and serves as a causative factor in patients with NSCLC.5,6 The resulting EML4–ALK fusion protein preserves the complete intracellular portion of ALK, and therefore, NSCLC cells with this fusion protein are highly sensitive to ALK tyrosine kinase inhibition, which could restrain tumor proliferation and induce tumor apoptosis.7 Crizotinib, a specific ALK inhibitor, is beneficial for most patients with ALK-positive NSCLC but has no obvious therapeutic effect on a minority of patients due to the acquired resistance to cerizotinib.8,9 It has been reported that the mutations in the ALK domain or the activated bypass signaling pathways contribute to crizotinib resistance in NSCLC.10–12
Cancer stem cells (CSCs), the aggressive subpopulation of cells within tumors, are responsible for tumorigenesis, relapse, and cancer metastasis associated with their capabilities of self-renewal and differentiation according to the CSC hypotheses.13–15 Numerous studies have documented the existence of CSCs in various types of human cancers including NSCLC.16,17 Additionally, emerging evidence has validated the clinical relevance of CSCs, including an initial positive response to therapy and resistance to currently used chemotherapy and radiotherapy.18 Chemotherapeutic agents are widely accepted as the standard therapy for patients with advanced NSCLC. Their effects are accompanied by chemotherapy resistance and multidrug resistance, which are possibly induced by the enrichment of CSCs.16
Genetic and epigenetic mechanisms are implicated in the malignant reprogramming process correlated with the acquisition and maintenance of self-renewal and stemness features of CSCs.19 It has been verified that ncRNAs, including miRNAs and lncRNAs, are involved in one such regulatory mechanism. For example, lncRNA HOTAIR and lncRNA MALAT1 play an important role in cancer metastasis.20,21 Of particular note is that long intergenic noncoding RNA, regulator of reprogramming (lincROR), located at chromosome 18q21.31, could reprogram differentiated cells to induced pluripotency stem cells and, therefore, is identified as a major regulator of pluripotency reprogramming.22,23 Moreover, Wang et al have shown the key role of lincROR in human embryonic stem cells as a potential self-renewal and pluripotency marker.24 Accumulating studies have reported the involvement of lincROR in various cancers, including lung adenocarcinoma,25 bladder cancer,22 pancreatic cancer,26 and breast cancer.27 Specially, lincROR is reported to be involved in chemoresistance in docetaxel-resistant lung adenocarcinoma cells.25 However, the potential roles of lincROR in the acquired resistance to crizotinib in NSCLC are not fully understood.
A previous study has demonstrated the association of stemness with EML4–ALK-driven tumorigenesis of NSCLC cells, verifying that the stem cell-like properties of NSCLC cells are increased both in vitro and in vivo by EML4–ALK.28 Furthermore, lincROR plays an important role in the reprogramming process relative to stemness features of CSCs. However, little is known about the association between EML–ALK and lincROR concerning stem cell-like properties of NSCLC cells.
In this study, we first investigated the expression profile and prognosis of lincROR in tissues and its association with ALK using quantitative polymerase chain reaction (qPCR) assay. Subsequently, we examined the potential role of ALK and lincROR in stem cell-like properties of EML–ALK+ cells. Additionally, the effects of crizotinib on lincROR expression were evaluated. Finally, we analyzed the role of lincROR in cells’ chemoresistance to crizotinib according to cell viability assay.
Patients and methods
Patients’ samples
Clinical tissues were collected from 106 patients who underwent a surgical procedure at the Zhongnan Hospital of Wuhan University (Wuhan, Hubei, China) between 2011 and 2016. No patient received therapy before surgical resection. Each patient had signed written informed consent prior to the study. The utilization of tumor samples was approved by the ethics committee of the Zhongnan Hospital of Wuhan University. Tumor tissues were immediately frozen in liquid nitrogen tanks at the time of resection until use. The clinicopathologic features of these samples are shown in Table 1.
Cell lines and cell culture
The cell line A549 was provided by the American Type Culture Collection (Manassas, VA, USA). The human EML–ALK+ NSCLC cell line H2228 was also obtained from the American Type Culture Collection. The human EML–ALK+ NSCLC cell line H3122 was purchased from Dana-Farber Cancer Institute (Boston, MA, USA). The cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum with 1% penicillin/streptomycin (Sigma-Aldrich Co., St Louis, MO, USA) in a humidified incubator with 5% CO2 at 37°C.
RNA extraction and real-time qPCR
Total RNA was extracted from the tissues or cells using the Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The RNA samples were reverse-transcribed into cDNA with the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Takara, Otsu, Japan). β-Actin was used as the internal control. All primers used for qPCR were obtained from GenePharma (Shanghai, China), and the sequences are listed in Table 2. qPCR was conducted with the SYBR PrimeScript qPCR kit (Takara) in a CFX Connect™ qPCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s instructions. All samples were analyzed in triplicate. Fold changes of ALK or lincROR were calculated by the relative quantification (2−ΔΔCt) method.
  | Table 2 The sequences used in this study |
ALK knockdown experiments
siRNA for ALK (ALK-si) and negative control (NC) were obtained from RiboBio (Guangzhou, China). The sequence of ALK-si is listed in Table 2. H3122 cell line was used for transfection with ALK-si. In brief, cells were planted into 12-well plates for 12 h and then transfected with ALK-si using X-treme GENE transfection reagent (Roche, Basle, Switzerland) according to the corresponding manufacturer’s recommendations. After 24 h transfection, total RNA was isolated and then used for analysis.
lincROR overexpression experiments
pcDNA-lincROR was constructed by introducing a BamHI-EcoRI fragment containing the lincROR DNA into the same site in pcDNA3.1. H3122 and H2228 cells were transfected with pcDNA-lincROR using Lipofectamine 2000 (Thermo Fisher Scientific) under the manufacturer’s protocol, respectively. RNA isolation was performed after 24 h transfection.
Cell counting kit-8 (CCK-8) assay
A colorimetric assay was used to analyze the cell viability of H3122 and H2228 cells using the CCK-8 reagent (Beyotime Biotechnology, Shanghai, China), respectively. Cells transfected with pcDNA-lincROR or pcDNA control were set as the experimental or the control group. After 24 h transfection, the cells were planted into 96-well plates supplemented with 10 μL of CCK-8 reagent and then incubated for 1 h at 37°C. Subsequently, the transfected cells were separately treated with crizotinib (TargetMol, Shanghai, China) at various concentrations (0, 0.25, 0.5, 1.0, and 5.0 μM) and then cultured for 24 h. Finally, the cell viability was analyzed by measuring the OD at 450 nm wavelength using the Tecan M200 multimode microplate reader (Tecan, Mechelen, Belgium).
Spheroid formation assay
Spheroid formation assay was performed to evaluate the capability of cell self-renewal using ultralow attachment surface 96-well culture plates. Cells transfected with ALK-si and pcDNA-lincROR, or ALK-si and pcDNA-control, or NC-si and pcDNA-control were, respectively, resuspended in 200 μL serum-free DMEM/F12 containing 20 ng/mL human epidermal growth factor, 20 ng/mL human basic fibroblast growth factor, and 1% nitrogen. The presence of spheroids was monitored after 10 days, and images of representative fields were captured by the microscope.
Flow cytometry analysis
During flow cytometry analysis, cells were dissociated as single cells and washed with PBS. Subsequently, these cells were resuspended in 1% bovine serum albumin with the corresponding control or specific antibody and then incubated for 30 min at room temperature. Finally, the expressions of CD133 and CD44 were measured by flow cytometry (BD Biotechnology, San Jose, CA, USA). Fluorescence-activated cell sorter was conducted to sort CD133 and CD44, respectively. CD133+CD44+, CD133+CD44−, CD133−CD44+, and CD133−CD44− subpopulations were obtained.
Statistical analyses
SPSS version 18.0 (SPSS, Chicago, IL, USA) was used to conduct statistical analysis. The unpaired t-test was used for analyzing the difference in two groups, and analysis of variance was used for multiple groups (>2). Chi-square test was applied for analyzing the classification variables between different groups. Kaplan–Meier and log-rank test were used to conduct survival analysis. All experiments mentioned previously were repeated three times. The results were presented as mean ± SD. P<0.05 was considered statistically significant in all statistical analyses.
Results
Upregulation of lincROR in EML–ALK+ NSCLC tissues and its relevance with prognosis of EML–ALK NSCLC patients
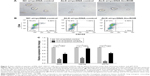
qPCR assay was performed to identify the expression levels of lincROR in EML–ALK NSCLC tissues. The clinicopathologic features of 106 patient samples are summarized in Table 1. lincROR expression was evidently associated with differentiation (P=0.0007), TNM stage (P=0.03212), lymph node metastasis (P=0.01956), distant metastasis (P=0.01304), and EML–ALK (P=0.02579). Ninety-five cases were identified as EML–ALK−, and the remaining (11 cases) were diagnosed as EML–ALK+. As shown in Figure 1A, lincROR expression in EML–ALK+ NSCLC tissues was significantly higher than that in EML–ALK− tumor tissues (P<0.01). The association between lincROR expression and prognosis was explored by Kaplan–Meier and log-rank tests. The median expression of lincROR in 68 EML–ALK NSCLC patients was defined as the cutoff to classify the patients into two groups (high expression and low expression). The progression-free survival of EML–ALK NSCLC patients was analyzed. A favorable prognosis was observed in low-lincROR expression patients with EML–ALK+ NSCLC (Figure 1B, P=0.027).
lincROR expression was decreased by ALK knockdown
The ALK knockdown assay was conducted to further investigate the relationship between lincROR and EML–ALK. The expressions of ALK and lincROR were measured by qPCR assay in H3122 cells transfected with ALK-si or NC-si. We found that lincROR expression was markedly restrained after the knockdown of ALK, indicating that lincROR expression may be associated with ALK (Figure 2, P<0.05).
Overexpression of lincROR rescued a part of stem cell-like properties of H3122 cells which were impaired by ALK knockdown
Sphere formation assay, flow cytometry analysis, and qPCR assay were used to explore the effects of ALK knockdown or lincROR overexpression on stem cell-like properties of H3122 cells. Sphere formation assay illustrated that the spheroid rate of H3122 cells transfected with ALK-si and pcDNA-control was lower than that of cells transfected with NC-si and pcDNA-control, but lincROR overexpression resulted in enhanced spheroid rate of H3122 cells transfected with ALK-si (Figure 3A). We observed that 6.857% (mean value, MD) CD133+CD44+ in NC-si + pcDNA-control group, 1.327% (mean value, MD) in ALK-si + pcDNA-control group, and 4.583% (mean value, MD) in ALK-si + pcDNA-lincROR group. The results indicated that ALK knockdown reduced the percentage of CD133+CD44+ in H3122 cells, but lincROR overexpression could partly improve the percentage of CD133+CD44+ in H3122 cells (Figure 3B). Results obtained with qPCR showed that Sox2, Nanog, and OCT4 expressions in ALK-si+pcDNA control group were all significantly lower than those in NC-si+pcDNA control group, indicating that ALK knockdown contributed to a decrease of Sox2, Nanog, and OCT4 expressions (Figure 3C, P<0.05). Conversely, Sox2, Nanog, and OCT4 expressions in ALK-si+pcDNA lincROR group were evidently higher than those in ALK-si+pcDNA control group, revealing that overexpression of lincROR upregulated the Sox2, Nanog, and OCT4 expressions when they were repressed by ALK knockdown (Figure 3C, P<0.05). Taken together, the results revealed that lincROR overexpression could enhance stemness features of EML–ALK+ NSCLC cells, which were repressed by ALK knockdown.
lincROR expression was inhibited by increased concentration of crizotinib in H2228 and H3122 cell lines
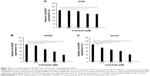
To determine the correlation of lincROR with crizotinib, we detected lincROR expression at different concentrations of crizotinib in A549, H2228, and H3122 cells, respectively. As displayed in Figure 4A, no obvious impact of crizotinib on lincROR expression was observed in A549 cells (P<0.05). However, remarkable downregulation of lincROR was detected in both H2228 and H3122 cells as a result of increased concentration of crizotinib (Figure 4B and C, P<0.05). These results implied that crizotinib could influence lincROR expression.
lincROR overexpression elevated chemoresistance to crizotinib in H3122 and H2228 cells
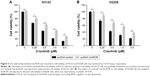
The effects of lincROR and crizotinib on cell viability of EML4–ALK+ NSCLC cells were explored. The results showed that the inhibitory effect of crizotinib on cell viability was increased with its increasing concentrations. The cell viability of H3122 cells was significantly higher in pcDNA-lincROR group as compared to pcDNA-control at various concentrations (0.25, 0.5, 1.0, and 5.0 μM) of crizotinib, respectively (Figure 5A, P<0.05). This was the case in H2228 cells (Figure 5B, P<0.05). It could be observed that lincROR overexpression elevated the chemoresistance to crizotinib in both H3122 and H2228 cells.
Discussion
EML4–ALK fusion has been identified as a causative factor in patients with NSCLC.29 lincROR, a key reprogramming regulator, has been found in different cancers where it acts as an oncogene.22,26,27 Especially in NSCLC, the oncogenic role and prognostic value of lincROR have been outlined as previously reported.30 However, little is known about the role of lincROR in EML–ALK-induced NSCLC. In this study, we first found elevated expression of lincROR in EML–ALK+ tissues in comparison with EML–ALK− tissues. Additionally, we found a favorable prognosis in low-expressed lincROR patients with EML–ALK+ NSCLC, in accordance with a previous finding.30 These results inspired us to speculate that lincROR might govern the tumorigenesis, progression, and prognosis of EML–ALK+ NSCLC.
Notably, EML–ALK is an activator for a variety of downstream signaling molecules such as STAT3, ERK, and AKT, which play a key role in inducing and maintaining stem-like properties for CSCs.28 It has been reported that CSCs have the capacity of increasing tumorigenesis and the potential of forming tumor metastasis. Furthermore, EML4–ALK has been discovered as the inducer for a stem-like phonotype in NSCLC cells.5,6,28 Major transcription factors, including OCT4, SOX2, and NANOG, are important for pluripotent stem cell phenotype.31 Of note, lincROR can be directly modulated by OCT4, SOX2, and NANOG.32 In addition, lincROR is involved in various cancers by acting as a tumor-promoting factor. It was, therefore, tempting to speculate some correlation between EML–ALK and lincROR in stem cell-like properties of NSCLC. In our work, lincROR exhibited downregulated expression in H3122 cells after transfection with ALK-si. We further discovered that stem cell-like properties of H3122 cells were downregulated after transfection with ALK-si. Interestingly, H3122 cells transfected with ALK-si displayed rescued stem cell-like properties induced by lincROR overexpression. Therefore, it is reasonable to presume that ALK could regulate the stemness features of EML–ALK+ NSCLC cells, possibly by modulating lincROR.
Crizotinib serves as the therapeutic drug for patients with EML–ALK+ NSCLC by directly targeting ALK and shows the anticancer activity in patients with ALK-positive NSCLC.33 Nevertheless, the occurrence of crizotinib resistance commonly leads to tumor recurrence and cancer metastasis.9 Pan et al have demonstrated that lincROR is associated with chemoresistance in NSCLC.25 To the best of our knowledge, the correlation between lincROR and crizotinib in EML–ALK+ NSCLC is less reported. Herein, we detected the expression pattern of lincROR in different concentrations of crizotinib. Our results indicated that lincROR expression was negatively correlated with the concentration of crizotinib. Moreover, we also found that lincROR could elevate cell viability of EML–ALK+ NSCLC cells, which was suppressed by crizotinib. These results indicated lincROR may promote acquired resistance to crizotinib in EML–ALK+ NSCLC.
In summary, this study provides a new insight into the important role of lincROR in the prognosis, stem cell-like properties, as well as acquired crizotinib resistance in EML–ALK+ NSCLC. However, this work also has some limitations. The functional role of lincROR in EML–ALK+ NSCLC and the concrete mechanism of lincROR in acquired crizotinib resistance were not explored. Further work should be conducted to gain a better understanding about the biological role of lincROR and how lincROR interacts with crizotinib in EML–ALK+ NSCLC.
Conclusion
This study reveals a key role of lincROR in the stemness features and the acquired crizotinib resistance associated with EML–ALK+ NSCLC, implying that lincROR could be a potential target in the prognosis and treatment of EML–ALK+ NSCLC. Further studies should concentrate on elucidating the exact molecular mechanisms of lincROR and crizotinib jointly acting on EML–ALK+ NSCLC.
Disclosure
The authors report no conflicts of interest in this work.
References
Stanek L, Springer D, Konopasek B, et al. Molecular pathological predictive diagnostics in a patient with nonsmall cell lung cancer treated with crizotinib therapy: a case report. Oncol Lett. 2017;14(6):7545–7548. | ||
Zhang YH, Gu XX, Jiang SJ, Xu LJ. Yangfei Kongliu Formula, a compound Chinese herbal medicine, combined with cisplatin, inhibits growth of lung cancer cells through transforming growth factor-β1 signaling pathway. J Int Med. 2017;15(3):242–251. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Peters S, Kerr KM, Stahel R. PD-1 blockade in advanced NSCLC: a focus on pembrolizumab. Cancer Treat Rev. 2017;62:39–49. | ||
Guo F, Liu X, Qing Q, et al. EML4-ALK induces epithelial-mesenchymal transition consistent with cancer stem cell properties in H1299 non-small cell lung cancer cells. Biochem Biophys Res Commun. 2015;459(3):398–404. | ||
Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7(9):1466–1476. | ||
Christensen JG, Zou HY, Arango ME, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6(12 Pt 1):3314–3322. | ||
Passaro A, Lazzari C, Karachaliou N, et al. Personalized treatment in advanced ALK-positive non-small cell lung cancer: from bench to clinical practice. Onco Targets Ther. 2016;9:6361–6376. | ||
Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4(6):662–673. | ||
Toyokawa G, Seto T. Updated evidence on the mechanisms of resistance to ALK inhibitors and strategies to overcome such resistance: clinical and preclinical data. Oncol Res Treat. 2015;38(6):291–298. | ||
Su J, Zhong W, Zhang X, et al. Molecular characteristics and clinical outcomes of EGFR exon 19 indel subtypes to EGFR TKIs in NSCLC patients. Oncotarget. 2017;8(67):111246–111257. | ||
Xiao H, Wang J, Liu Y, Li L. Relative influence of c-Kit expression and epidermal growth factor receptor gene amplification on survival in patients with non-small cell lung cancer. Oncology Lett. 2014;8(2):582–588. | ||
Wang X, Zhu Y, Ma Y, et al. The role of cancer stem cells in cancer metastasis: new perspective and progress. Cancer Epidemiol. 2013;37(1):60–63. | ||
Fessler E, Dijkgraaf FE, De Sousa E, Melo F, Medema JP. Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett. 2013;341(1):97–104. | ||
Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. Embo Rep. 2014;15(3):244–253. | ||
Wang L, Liu X, Ren Y, et al. Cisplatin-enriching cancer stem cells confer multidrug resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis. 2017;8(4):e2746. | ||
Chen L, Long C, Tran KAM, Lee J. A synthetic binder of breast cancer stem cells. Chemistry. 2018;24(15):3694–3698. | ||
Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int J Biochem Cell Biol. 2012;44(12):2144–2151. | ||
Muñoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012;6(6):620–636. | ||
Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7(1):90. | ||
Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. | ||
Chen Y, Peng Y, Xu Z, et al. LncROR promotes bladder cancer cell proliferation, migration, and epithelial-mesenchymal transition. Cell Physiol Biochem. 2017;41(6):2399–2410. | ||
Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Database issue):D146–D151. | ||
Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. | ||
Pan Y, Chen J, Tao L, et al. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget. 2017;8(20):33144–33158. | ||
Zhan HX, Wang Y, Li C, et al. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374(2):261–271. | ||
Hou P, Zhao Y, Li Z, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5(5):e1287. | ||
Oh SJ, Noh KH, Lee YH, et al. Targeting stemness is an effective strategy to control EML4-ALK+ non-small cell lung cancer cells. Oncotarget. 2015;6(37):40255–40267. | ||
Pyo KH, Sun ML, Kim HR, et al. Establishment of a conditional transgenic mouse model recapitulating EML4-ALK-positive human non-small cell lung cancer. J Thorac Oncol. 2016;12(3):491–500. | ||
Qu CH, Sun QY, Zhang FM, Jia YM. Long non-coding RNA ROR is a novel prognosis factor associated with non-small-cell lung cancer progression. Eur Rev Med Pharmacol Sci. 2017;21(18):4087–4091. | ||
Ding DF, Li XF, Xu H, et al. Mechanism of resveratrol on the promotion of induced pluripotent stem cells. J Integr Med. 2013;11(6):389–396. | ||
Wang S, Liu F, Deng J, Cai X, Han J, Liu Q. Long noncoding RNA ROR regulates proliferation, invasion, and stemness of gastric cancer stem cell. Cell Reprogram. 2016;18(5):319–326. | ||
Alshareef A, Gupta N, Zhang HF, Wu C, Haque M, Lai R. High expression of β-catenin contributes to the crizotinib resistant phenotype in the stem-like cell population in neuroblastoma. Sci Rep. 2017;7(1):16863. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.