Back to Journals » OncoTargets and Therapy » Volume 13
Linc01094 Accelerates the Growth and Metastatic-Related Traits of Glioblastoma by Sponging miR-126-5p
Received 20 May 2020
Accepted for publication 1 August 2020
Published 6 October 2020 Volume 2020:13 Pages 9917—9928
DOI https://doi.org/10.2147/OTT.S263091
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xin Xing Li, Qi Yu
Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang 110004, Liaoning, People’s Republic of China
Correspondence: Qi Yu
Department of Neurosurgery, Shengjing Hospital of China Medical University, No. 36 Sanhao Street, Heping District, Shenyang 110004, Liaoning, People’s Republic of China
Email [email protected]
Background: Long intergenic non-coding RNAs (lincRNAs) are associated with the progression of glioblastoma (GBM). However, how linc01094 contributes to the growth and metastatic phenotypes of GBM remains not fully studied.
Methods: The expression levels of linc01094 and miR-126-5p in GBM tissues and cell lines were analyzed using qRT-PCR. Loss-of-function experiments were performed to detect the biological activity of linc01094 in GBM. Glioblastoma tumor model was constructed to explore the impact of linc01094 on GBM cell growth in vivo. Linc01094-sponged miR-126-5p was certified by luciferase reporter assay and RNA immunoprecipitation (RIP). The protein expression of miRNA target gene, dynactin subunit 4 (DCTN4) was detected using Western blotting assay.
Results: Herein, we observed that the level of linc01094 was higher in GBM tissues. Silencing of linc01094 restrained the growth and invasive abilities of GBM cell. Moreover, linc01094 level was negatively associated with miR-126-5p level in GBM and linc01094 acted as a “sponge” for miR-126-5p. Reintroduction of linc01094 reversed the tumor-inhibiting effects of miR-126-5p in GBM.
Conclusion: Altogether, linc01094 promoted the tumorigenesis and metastatic phenotypes of GBM cell by modulating of miR-1126-5p/DCTN4 signaling axis.
Keywords: linc01094, GBM, miR-126-5p, DCTN4
Introduction
Glioma is one of the most common types of primary brain tumors and can be classified into four grades (I, II, III and IV). High-grade glioma is identified as grades III and IV while low-grade glioma (LGG) belongs to grades I and II. Glioblastoma (GBM) belongs to the IV glioma.1,2 The cross-talk between microbiota and brain may also have crucial impacts during basic neurogenerative processes, in neurodegenerative disorders and tumors of central nervous system.3,4 Although diverse therapeutic options including surgical excision, radiotherapy and chemotherapy have been developed, the heterogeneity of GBM makes the cancer cell be less sensitive to chemo/radiotherapy.5,6 Meanwhile, the aggressive characteristics of GBM cell lead to the high recurrence rate and poor clinical outcomes of this disease.7 Therefore, the clinical outcomes of the current therapies for patients with GBM are unsatisfying and the overall survival (OS) prognosis is poor. Thus, it is urgent to develop novel therapies for the treatment of GBM.
LncRNAs are a new kind of non-protein coding RNAs with a length of more than 200 nt.8 Recently, a series of lncRNAs have been demonstrated to be tumor markers in GBM.9,10 For example, lncRNA PCED1B Antisense RNA 1 (PCED1B-AS1) promotes the Warburg effect and tumorigenesis via upregulating hypoxia inducible factor 1 subunit alpha (HIF-1α) in GBM.11 LncRNA ADAMTS9 Antisense RNA 2 (ADAMTS9-AS2) promotes Temozolomide (TMZ) resistance by intervening the FUS/MDM2 axis in GBM cell.12 Competing endogenous RNAs (ceRNAs) are important in elucidating how lncRNAs serve as scaffolds with microRNAs (miRNA). LncRNA FEZF1 Antisense RNA 1 (FEZF1-AS1) might sponge miR-34a to modulate Notch receptor 1 (Notch-1) in GBM, thereby accelerating cancer cell migration and invasion.13 UBE2R2 Antisense RNA 1 (UBE2R2-AS1) induces glioma cell apoptosis through intervening the miR-877-3p/TLR4 axis.14 A recent study has found that linc01094 is upregulated in clear cell renal cell carcinoma (ccRCC) and acts as an oncogene in ccRCC via regulating miR-224-5p/CHSY1.15 Nevertheless, the precise functional roles of linc01094 in GBM are still unknown.
miRNA are non-coding RNAs derived from endogenous chromosomes that consist of approximately 22 nucleotides. Recently, a variety of miRNAs is expressed in an abnormal fashion and function as oncogenes or tumor suppressors in cancers. In ovarian cancer, miR-126-3p inhibits the proliferation and invasion of cancer cells through targeting Plexin B2 (PLXNB2).16 MiR-126 has also been reported to be downregulated in colon cancer and controls tumor cell growth, metastasis and survival.17 In human glioma, miR-126 regulates the ERK pathway via targeting KRAS to inhibit glioma cell growth and invasion.18 Xu et al, demonstrates that miR-126 affects the invasion and migration of glioma cells through targeting GATA Binding Protein 4 (GATA4).19 Dynactin is a multiple-subunit protein compound involved in the activation of most forms of cytoplasmic dynein in eukaryotes. Six subunits of dynactin, DCTN1, DCTN2, DCTN3, DCTN4, DCTN5, and DCTN6, have been verified, and all the subunits are encoded through their corresponding genes.20 In colon cancer, higher expression level of DCTN4 is significantly correlated with the satisfactory overall survival of patients.21 Nevertheless, after bibliographic retrieval, the correlation between miR-126 and linc01094 in glioma has not been completely elucidated.
In the current study, the differentially expressed lincRNAs in GBM tissues and normal tissues were identified using GEO dataset and The Cancer Genome Atlas (TCGA) database. linc01094 was upregulated in GBM and selected as the object of our current study. We investigated the biological role of linc01094 in human GBM. Loss function assays indicated that downregulation of linc01094 inhibited GBM cell growth and invasion ability, indicating the oncogenic role of linc01094 in GBM. Importantly, we revealed that linc01094 acted as a “sponge” for miR-126-5p and regulated its target gene, DCTN4.
Materials and Methods
Cell Lines and GBM Tissues
U87MG and U251 cell lines were purchased from Procell Life Science&Technology Co., Ltd (Wuhan, Hubei, China). U251 cell was cultured in DMEM and U87MG cell was cultured in MEM supplement with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA). A total of 40 GBM specimens and 40 non-neoplastic brain tissues from epileptogenic patients were collected from Shengjing Hospital of China Medical University. Written informed consent was obtained from all participants who were involved in the study. This research was approved by the Ethics Committee of the Shengjing Hospital of China Medical University (approval No. SJ-2015-052). The clinicopathological features of all patients are summarized in Table 1.
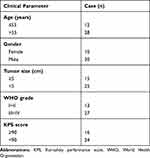 |
Table 1 The Clinicopathologic Features in Patients with GBM |
Selection of Microarrays in Gene Expression Omnibus (GEO)
GBM-related microarray (GSE15824) was downloaded from the National Center of Biotechnology Information (NCBI) GEO (http://www.ncbi.nlm.nih.gov/geo/) database. Fold change >1 or <-1 and P-value < 0.01 between the GBM and normal brain tissues were considered as significant. The expression level of the desired RNA was normalized by the Deseq package of the R language for further analysis.
Target miRNAs Prediction
Two prediction algorithms, miRDB (http://www.mirdb.org/custom.html) and LncBase Predicted v.2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-experimental) were used to predict the miRNAs that interact with linc01094. Venn diagrams were used to identify overlapping target miRNAs (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Cell Transfections
The shRNAs targeted linc01094 (designated as sh-linc01094 #1 and sh-linc01094 #2) were synthesized by RiboBio (Guangdong, Guangzhou, China), and shRNA scrambled was used as negative control (sh-NC). The pcDNA3.1 carrying linc01094 overexpression vector (pcDNA3.1-linc01094), pcDNA3.1 carrying DCTN4 overexpression vector (pcDNA3.1-linc01094) and negative pcDNA3.1 vector were purchased from GenePharma (Shanghai, China). miR-126-5p mimics and miRNA negative control (miR-NC) were purchased from Ribobio. U87MG and U251 cell transfections were conducted using Lipofectamine 3000 (Thermo Fisher Scientific). The sh-linc01094 #1 or sh-NC were packaged into lentivirus and infected into U87MG cell to obstruct linc01094 stable knockdown cell line.
Cell Proliferation
U87MG or U251 cells (2 × 103) were plated into 96-well plates. After transfection, 10 μL of cell counting kit-8 (CCK-8) solution was added to 96-well plates at 0 h, 24 h, 48 h, 72 h, or 96 h, respectively. After incubated for 2 h in incubator, the OD value was detected at 450 nm. Cell proliferation was also detected using 5-ethynyl-2ʹ-deoxyuridine (EdU) incorporation test with an EdU assay kit (Life Technologies Corporation, Carlsbad, CA, USA) according to the instructions.
Colony Formation Assay
U251 or U87MG cells (1× 103) were cultured into 6-well plates. The medium in plate was changed every three days. After two weeks, cell colonies were dyed with 1% crystal violet (Sigma, Shanghai, China).
Invasion Analysis
Cells (2 × 104) were cultured into the upper chamber of transwell filter (BD Biosciences, Franklin Lakes, NJ, USA) with an Matrigel-coated membrane. Seven hundred microliter of complete medium supplement with 20% FBS was added into the lower chamber. After incubation for 24 hours, the number of invading cells on the lower surface was counted under an inverted microscope.
Luciferase Reporter Assay
The fragment of the 3ʹ-untranslated region (3ʹ-UTR) of DCTN4 that containing the wild-type (wt) or mutated type (mut) miR-126-5p pairing sequences, the fragment of linc01094 that containing the wild-type (wt) or mutated type (mut) miR-126-5p pairing sequences were provided by GenePharma. The sequences of DCTN4 3ʹ-UTR (DCTN4-wt or DCTN4-mut) were cloned into pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The sequences of linc01094 3ʹ-UTR (linc01094-wt or linc01094-mut) were cloned into pGL3 luciferase reporter vector. U87MG or U251 cells (5 × 105) were cultured in 24 well plates and cotransfected with reporter plasmid and miR-126-5p mimics. After 48 hours transfection, the relative luciferase activity was measured.
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted from tissues or cells using TRIzol reagent (Thermo Fisher Scientific). qRT-PCR was performed using a SYBR Green PCR Kit (Takara, Dalian, China) on a CFX96 Touch sequence detection system (Bio-Rad, USA). GAPDH was the internal control. For miRNA quantitative assay, miRNA was extracted using the miRNeasy Mini Kit (Qiagen, Germantown, MD, USA). The level of miR-126-5p was detected using an All-in-One™ miRNA qPCR Detection kit. The snRNA U6 was used as the internal control. The levels of genes expression were calculated using the 2−ΔΔCT method. The primer sequences were as follows: linc01094: Forward, 5′-TGTAAAACGACGGCCAGT-3′ and Reverse, 5′-CAGGAAACAGCTATGACC-3′; miR-126-5p: Forward, 5′-GCCGGCGCCCGAGCTCTGGCTC-3′ and Reverse, 5′-CATTATTACTTTTGGTACGCG-3′; DCTN4: Forward, 5′-CACACCCTCTCTACTCGGG-3′ and Reverse, 5′-ACATGCCAGGTAATAGGCTTTC-3′; GAPDH: Forward, 5′-AATGGATTTGGACGCATTGGT-3′ and Reverse, 5′-TTTGCACTGGTACGTGTTGAT-3′; U6: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and Reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Immunoblotting
Proteins were extracted from cells using RIPA lysis buffer (Beyotime Biotechnology, Nanjing, China). Equal aliquots of lysates were separated by 8% SDS-PAGE gel and then transferred to PVDF membrane (Millipore, Braunschweig, Germany). Then membrane was blocked with 5% blocking solution for 1.5 h, followed by incubation with DCTN4 or GAPDH antibody (1:1000, Proteintech Group, Inc, Rosemont, IL, USA) overnight at 4°C. Finally, membranes were incubated with HRP-conjugated secondary antibody (1:10,000, Beyotime Biotechnology) for 2 hours. The bands were visualized by ECL detection system (Millipore).
RNA Immunoprecipitation (RIP)
Ago2-RIP assay was conducted in U251 and U87MG cell using Ago2 antibody or normal IgG-conjugated Protein A resin (Sigma-Aldrich, USA). U87MG or U251 cell lysis was incubated with magnetic beads that coated with antibody against Ago2 or immunoglobulin G for 4 hours at 4°C. The abundances of linco01094 or miR-126-5p in the precipitates were determined by qRT-PCR.
Fluorescence in situ Hybridization (FISH)
RNA FISH probe specifically designed for linc01994 or miR-126-5p was purchased from RiboBio (Guangdong, Guangzhou, China). U87MG or U251 cells were air-dried and incubated with FISH probe in hybridization buffer. After DAPI staining for nuclear, fluorescence signal was detected using fluorescence microscope (Olympus).
Isolation of Cytoplasmic-Nuclear RNA
U251 and U87MG cell lines were rinsed twice in PBS. Cell suspension in lysis buffer was incubated on ice for about 20 minutes and then centrifuged for 10 seconds at 13,000 rpm. Cytoplasmic RNA was in the supernatant. After centrifugation for 15 minutes, the nuclear pellet was obtained in lysis buffer and incubated on ice for 20 minutes, followed by centrifugation to discard the insoluble material. The qRT-PCR was used to determine linc01094 expression in the nucleus or cytoplasm, normalizing to U6 or GAPDH.
Tumor Xenograft Model
After sh-linc01094 or sh-NC stable transfection, 100 μL of U87MG cells (2×107) were subcutaneously implanted into the back of BALB/c nude mice (3 mice in each group). The length and width of xenograft tumors were measured every week and tumor volume was calculated according to the formula: (length x width2)/2. After 21 days, the nude mice were sacrificed. Xenograft tumors were subjected for immunohistochemical (IHC) staining. Animal experiment was approved by the Ethics Committee of Shengjing Hospital of China Medical University (approval No. 2018–0912) and was compiled with the Guide for the Care and Use of Laboratory Animals (NIH publication No. 86–23, revised 1985).
Statistical Analysis
Statistical analysis was completed using GraphPad version 7.0. The relationship between linc01094 and miR-126-5p or DCTN4 was determined using Spearman correlation analysis. The difference was evaluated by Student’s t-test or one-way analysis of variance post hoc Tukey’s test, and P<0.05 is statistically significant.
Results
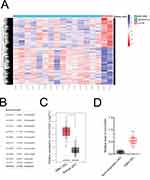
Linc01094 is Upregulated in GBM
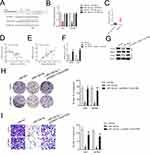
To seek the dysregulated lincRNAs in GBM, we conducted a comprehensive gene profiling analysis using the GEO dataset (GSE15824). Through analysis of microarray data, the expression of linc01094 was found to be the most upregulated lincRNA in GBM (Figure 1A and B). We further evaluated the expression level of linc01094 using the RNA-Seq datasets GEPIA (http://gepia.cancer-pku.cn/index.html). As shown in Figure 1C, linc01094 was remarkedly upregulated in GBM tissue vs normal tissue. To verify the results, we analyzed the expression levels of linc01094 in GBM samples and non-neoplastic tissues by qRT-PCR assay, and linc01094 was found to be upregulated in GBM tissues when compared to non-neoplastic tissues (Figure 1D).
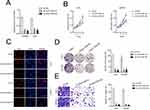
Linc01094 Silencing Suppresses the Growth and Invasion Ability of GBM Cell
U251 and U87MG cell lines were firstly transfected with sh-linc01094 #1 or sh-linc01094 #2 and the transfection efficiency of linc01094 was tested by qRT-PCR assay (Figure 2A). The impacts of linc01094 on the proliferation of sh-linc01094 transfected cells were measured using CCK-8 and Edu assay. As indicated in Figure 2B and C, linc01094 silencing considerably repressed the cell viability of U87MG and U251 in vitro. Consistently, the colony forming capacities of U87 and U251 cell were also impaired by sh-linc01094 as demonstrated by colony formation assay (Figure 2D). In transwell invasion test, we observed that downregulation of linc01094 decreased the invasive potential in U87MG and U251 cell (Figure 2E).
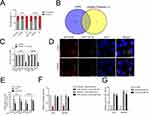
miR-126-5p Displays a Negative Correlation with Linc01094
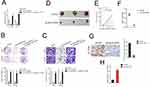
Then, cytoplasm and nuclear fractions assay suggested that linc01094 was mainly localized in the cytoplasm but not nucleus (Figure 3A). To illustrate the mechanism underlying which linc01094 contributes to GBM, bioinformatics tools (miRDB and LncBase Predicted v.2) were used to screen miRNAs that have the hypothetic binding sites within linc01094 (Figure 3B). The levels of these miRNAs in U87MG and U251 cell were detected using qRT-PCR. We found that miR-126-5p rather than other miRNAs exhibited a decreased expression in both pcDNA3.1-linc01094 transfected U87MG and U251 cell compared with in pcDNA3.1 group (Figure 3C). The following result of RNA-FISH test validated the colocalization between fractions of linc01094 and miR-126-5p (Figure 3D). Moreover, the result from RIP assay indicated that linc01094 and miR-30a-5p were enriched in Ago2 pellet as compared with in the input control (Figure 3E). The following luciferase reporter experiment indicated that miR-126-5p mimics restrained the relative luciferase activity of the report vector containing linc01094-wt 3′-UTR rather than linc01094-mut 3′-UTR (Figure 3F). Finally, we checked the level of miR-126-5p in GBM cell after sh-linc01094 transfection using qRT-PCR. As shown in Figure 3G, the level of miR-126-5p was significantly raised in sh-inc01094 groups in comparison with sh-NC group. All these observations indicated that linc01094 negatively regulated the expression of miR-126-5p.
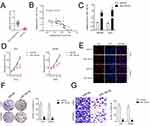
miR-126-5p Suppresses GBM Cell Proliferation and Invasion
To study the roles of miR-126-5p in GBM, we compared the levels of miR-126-5p in 40 paired of GBM tissues and non-neoplastic tissues. In Figure 4A, miR-126-5p was downregulated in GBM samples. Pearson correlation analysis shown that miR-126-5p level was negatively associated with linc01094 expression in GBM (Figure 4B). U87MG and U251 cell lines were transfected with miR-126-5p mimics and the transfection efficiency was checked by qRT-PCR assay (Figure 4C). The impacts of miR-126-5p on U87MG and U251 cell proliferation were detected by CCK-8 and Edu assay. As shown in Figure 4D and E, miR-126-5p significantly inhibited the cell proliferation of U87MG and U251 cell in vitro. Consistently, the colony forming in U87MG and U251 cell were also noticeably weakened by miR-126-5p as demonstrated by colony formation assay (Figure 4F). In transwell invasion test, we observed that upregulation of miR-126-5p decreased the invasive abilities of U87MG and U251 cell (Figure 4G).
DCTN4 is a Target of miR-126-5p
Bioinformatics tool RNA22 was used to predict the downstream target of miR-126-5p and we identified that DCTN4 had the putative binding targets between miR-126-5p (Figure 5A). The following luciferase reporter assay indicated that miR-126-5p mimics restrained the luciferase activity of the reporting vector containing DCTN4-wt 3′-UTR rather than DCTN4-mut 3′-UTR luciferase reporter vector (Figure 5B). Next, qRT-PCR result had shown that DCTN4 was upregulated in GBM tissues compared with in non-neoplastic tissues (Figure 5C). Pearson correlation analysis showed that DCTN4 had a significantly negative correlation with miR-126-5p and positively correlation with linc01094 in GBM tissues (Figure 5D and E). qRT-PCR assay and immunoblotting analysis also suggested that miR-126-5p mimics weakened the level of DCTN4 whereas cotransfection with pcDNA3.1-linc01094 neutralized the suppressive effect of miR-126-5p on DCTN4 expression (Figure 5F and G). Next, the colony formation and invasion assays were conducted to determine whether linc01094 serves as a sponge for miR-126-5p. As shown in Figure 5H and I, pcDNA3.1-linc01094 reversed the proliferative viability and invasion ability of GBM cell that were impaired by miR-126-5p.
Linc01094 Impedes Xenograft Tumor Growth of GBM Cell in vivo
Rescue experiments were carried out to determine whether overexpression of DCTN4 neutralized the effect of sh-linc01094 on GBM cell growth and invasion. pcDNA3.1 carrying DCTN4 overexpression vector (pcDNA3.1-DCTN4) was transfected into GBM cells which had been transfected with sh-linc01094 #1. Colony formation assay showed that the weakened growth induced by sh-linc01094 in GBM cells was abrogated by the introduction of pcDNA3.1-DCTN4 (Figure 6B). Consistently, transwell invasion assay displayed that the invasion of GBM cells inhibited by sh-linc01094 in was rescued by pcDNA3.1-DCTN4 (Figure 6C). Finally, a xenograft tumor model using U87MG cell was constructed to analysis the growth inhibition effect of sh-linc01094. As shown in Figure 6D–F, the tumor weight and tumor volume in sh-linc01094 group were lowered when compared with these in sh-Con group. IHC staining assay using tumor tissues displayed that the expression level of DCTN4 was decreased in sh-linc01094 group (Figure 6G). Nevertheless, the expression level of miR-126-5p was elevated in sh-linc01094 group compared with that in sh-Con group as demonstrated by qRT-PCR assay (Figure 6H).
Discussion
Glioma has been regarded as a kind of aggressive malignancy of the central nervous system worldwide.22 GBM is one of the most common malignant primary brain tumors. The five-year survival rate of patients with GBM is unfavorable, less than 5%.23,24 A deeper exploration of the pathogenesis of GBM is helpful for developing new treatment approaches and improving the clinical outcomes of patients. In our study, we proved that the GBM-related lncRNA, linc01094, was considerably upregulated in GBM. Consistently, using the GEPIA tool, we found that linc01094 was upregulated in TCGA GBM cohort.25 Through CCK-8 proliferation assay, EdU assay and colony formation assay as well as invasion analysis, we also found that linc01094 silencing inhibits the growth, colony formation and invasiveness of GBM cell in vitro.
Previous reports have certified that the molecular mechanism of lncRNAs is associated with its cellular localization.26,27 LncRNAs located in cytoplasm can function as miRNAs “sponge” to modulate the expressions of miRNA target genes by competition for shared miRNA response elements.28 Increasing investigations reveal a ceRNA mechanism regulated by lncRNAs in diverse malignancies.29 In the current study, cytoplasm and nuclear fractions test uncovered that linc01094 was primarily localized in cytoplasm, so we hypothesized that linc01094 might function as a ceRNA. In addition, RNA-FISH and RIP assays demonstrated the direct interaction between linc01094 and miR-126-5p. Meanwhile, level of miR-126-5p was negatively regulated by linc01094. All these data indicated that linc01094 severed as a “sponge” for miR-126-5p in GBM cell.
An increasing number of reporters have disclosed that miR-126-5p is relevant with diverse biological process, including cancer cell growth, apoptosis, metastasis and chemotherapy-resistant.30–32 For instance, miR-126-5p is downregulated in cervical carcinoma and upregulation of miR-126-5p induces cervical cancer cell apoptosis via targeting B-Cell CLL/Lymphoma 2 (Bcl-2).30 miR-126-5p is lowly expressed in colon cancer and YAP1-induced metastasis associated lung adenocarcinoma transcript 1 (MALAT1) promotes epithelial-mesenchymal transition (EMT) and angiogenesis by sponging miR-126-5p in colorectal cancer.31 In accordance, we uncovered that miR-126-5p was significantly downregulated in GBM tissues, and its level was negatively relevant with linc01094. MiRNAs are a kind of non-coding RNAs that regulate the expressions of their target genes at the post-transcriptional level through binding to their 3ʹ-UTRs.33 DCTN4 gene was identified as a target of miR-126-5p by luciferase reporter gene and the expression of DCTN4 was negatively modulated by miR-126-5p in GBM cell.
To better understand the regulatory axis of linc01094-miR-126-5p in human GBM, we constructed GBM cell line that was cotransfected with miR-126-5p and linc01094. The expression of DCTN4 impaired by miR-126-5p was offset by licn01094 cotransfection. Consistently, we showed that linc01094 overexpression canceled the GBM progression suppressed by transfection of miR-126-5p, indicating that linc01094 mediated GBM cell aggressive traits via regulating miR-126-5p. Altogether, our present study disclosed that linc01094 served as a sponge for miR-126-5p to decline the suppressive impact of miR-126-5p on DCTN4, and thus promoted the progression of GBM.
Funding
This work was supported by “Silybin Induced Autophagy of Glioma Stem Cells to Play An Anti-Tumor Molecular Mechanism” of Key Research and Development Project in Liaoning Province [Grant Number: 2018225094].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhang L, Wang Q, Wang F, et al. LncRNA LINC01446 promotes glioblastoma progression by modulating miR-489-3p/TPT1 axis. Biochem Biophys Res Commun. 2018;503(3):1484–1490. doi:10.1016/j.bbrc.2018.07.067
2. Jiang G, Dong H, Dong Y, Yang X. Long noncoding RNA unigene56159 promotes glioblastoma multiforme cell proliferation and invasion through negatively regulating microRNA1945p. Mol Med Rep. 2020;21(2):768–776. doi:10.3892/mmr.2019.10852
3. Ma Q, Xing C, Long W, et al. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. 2019;16(1):53. doi:10.1186/s12974-019-1434-3
4. Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin Immunol. 2017;32:25–34. doi:10.1016/j.smim.2017.08.001
5. Gao Y, Xu Y, Wang J, et al. lncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol Res. 2019;27(3):341–347. doi:10.3727/096504018X15228909735079
6. Wang S, Guo X, Lv W, et al. LncRNA RPSAP52 upregulates TGF-beta1 to increase cancer cell stemness and predict postoperative survival in glioblastoma. Cancer Manag Res. 2020;12:2541–2547. doi:10.2147/CMAR.S227496
7. Mazor G, Levin L, Picard D, et al. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 2019;10(3):246. doi:10.1038/s41419-019-1477-5
8. Li Z, Zhang J, Zheng H, et al. Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. J Exp Clin Cancer Res. 2019;38(1):380. doi:10.1186/s13046-019-1371-0
9. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37(1):265. doi:10.1186/s13046-018-0941-x
10. Wang H, Li L, Yin L. Silencing LncRNA LOXL1-AS1 attenuates mesenchymal characteristics of glioblastoma via NF-kappaB pathway. Biochem Biophys Res Commun. 2018;500(2):518–524. doi:10.1016/j.bbrc.2018.04.133
11. Yao Z, Zhang Q, Guo F, et al. Long noncoding RNA PCED1B-AS1 promotes the warburg effect and tumorigenesis by upregulating HIF-1alpha in glioblastoma. Cell Transplant. 2020;29:963689720906777. doi:10.1177/0963689720906777
12. Yan Y, Xu Z, Chen X, et al. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination axis. Front Cell Dev Biol. 2019;7:217. doi:10.3389/fcell.2019.00217
13. Luo L, Zhang Y, He H, et al. LncRNA FEZF1-AS1 sponges miR-34a to upregulate notch-1 in glioblastoma. Cancer Manag Res. 2020;12:1827–1833. doi:10.2147/CMAR.S240531
14. Xu W, Hu GQ, Da Costa C, et al. Long noncoding RNA UBE2R2-AS1 promotes glioma cell apoptosis via targeting the miR-877-3p/TLR4 axis. Onco Targets Ther. 2019;12:3467–3480. doi:10.2147/OTT.S201732
15. Jiang Y, Zhang H, Li W, et al. FOXM1-activated LINC01094 promotes clear cell renal cell carcinoma development via MicroRNA 224-5p/CHSY1. Mol Cell Biol. 2020;40(3):e00357–19. doi:10.1128/MCB.00357-19
16. Xiang G, Cheng Y. MiR-126-3p inhibits ovarian cancer proliferation and invasion via targeting PLXNB2. Reprod Biol. 2018;18(3):218–224. doi:10.1016/j.repbio.2018.07.005
17. Huang W, Lin J, Zhang H. miR-126: a novel regulator in colon cancer. Biomed Rep. 2016;4(2):131–134. doi:10.3892/br.2015.549
18. Li Y, Li Y, Ge P, Ma C. MiR-126 regulates the ERK pathway via targeting KRAS to inhibit the glioma cell proliferation and invasion. Mol Neurobiol. 2017;54(1):137–145. doi:10.1007/s12035-015-9654-8
19. Xu Y, Xu W, Lu T, Dai Y, Liang W. miR-126 affects the invasion and migration of glioma cells through GATA4. Artif Cells Nanomed Biotechnol. 2017;45(6):1–7. doi:10.1080/21691401.2017.1396222
20. Cianfrocco MA, DeSantis ME, Leschziner AE, Reck-Peterson SL. Mechanism and regulation of cytoplasmic dynein. Annu Rev Cell Dev Biol. 2015;31(1):83–108. doi:10.1146/annurev-cellbio-100814-125438
21. Wang S, Wang Q, Zhang X, et al. Distinct prognostic value of dynactin subunit 4 (DCTN4) and diagnostic value of DCTN1, DCTN2, and DCTN4 in colon adenocarcinoma. Cancer Manag Res. 2018;10:5807–5824. doi:10.2147/CMAR.S183062
22. Li M, Long S, Hu J, et al. Systematic identification of lncRNA-based prognostic biomarkers for glioblastoma. Aging (Albany NY). 2019;11(21):9405–9423. doi:10.18632/aging.102393
23. Tian Y, Zheng Y, Dong X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem. 2019;120(6):9056–9062. doi:10.1002/jcb.28180
24. Liao K, Lin Y, Gao W, et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol Ther Nucleic Acids. 2019;18:388–399. doi:10.1016/j.omtn.2019.09.005
25. Yang M, Zhai Z, Guo S, et al. Long non-coding RNA FLJ33360 participates in ovarian cancer progression by sponging miR-30b-3p. Onco Targets Ther. 2019;12:4469–4480. doi:10.2147/OTT.S205622
26. Liu G, Pan Y, Li Y, Xu H. lncRNA and mRNA signature for prognosis prediction of glioblastoma. Future Oncol. 2020;16(13):837–848. doi:10.2217/fon-2019-0538
27. Yan Y, Zhang L, Jiang Y, et al. LncRNA and mRNA interaction study based on transcriptome profiles reveals potential core genes in the pathogenesis of human glioblastoma multiforme. J Cancer Res Clin Oncol. 2015;141(5):827–838. doi:10.1007/s00432-014-1861-6
28. Chen D, Fan Y, Wan F. LncRNA IGBP1-AS1/miR-24-1/ZIC3 loop regulates the proliferation and invasion ability in breast cancer. Cancer Cell Int. 2020;20(1):153. doi:10.1186/s12935-020-01214-x
29. Hu T, Wang F, Han G. LncRNA PSMB8-AS1 acts as ceRNA of miR-22-3p to regulate DDIT4 expression in glioblastoma. Neurosci Lett. 2020;728:134896. doi:10.1016/j.neulet.2020.134896
30. Wang C, Zhou B, Liu M, Liu Y, Gao R. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25(4):463–470. doi:10.3727/096504016X14685034103879
31. Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38(14):2627–2644. doi:10.1038/s41388-018-0628-y
32. Shi H, Bi H, Sun X, et al. Tubeimoside-1 inhibits the proliferation and metastasis by promoting miR-126-5p expression in non-small cell lung cancer cells. Oncol Lett. 2018;16(3):3126–3134. doi:10.3892/ol.2018.9051
33. Ji Q, Xu X, Song Q, et al. miR-223-3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther. 2018;26(5):1299–1312. doi:10.1016/j.ymthe.2018.03.009
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.