Back to Journals » OncoTargets and Therapy » Volume 12
LINC00339 promotes growth and invasiveness of hepatocellular carcinoma by the miR-1182/SKA1 pathway
Received 2 March 2019
Accepted for publication 1 May 2019
Published 6 June 2019 Volume 2019:12 Pages 4481—4488
DOI https://doi.org/10.2147/OTT.S207397
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Jun Xiao, Haibo Yu, Zhongwu Ma
Department of Hepatobiliary Surgery, Wenzhou Central Hospital, Wenzhou 325000, People’s Republic of China
Background: Extensive research has shown that long noncoding RNA (lncRNA) is involved in tumorigenesis, including hepatocellular carcinoma (HCC). The lncRNA LINC00339 was reported to regulate the development of lung cancer or breast cancer. However, whether LINC00339 participates in HCC progression remains unclear. Here, our results showed that LINC00339 was upregulated in HCC.
Methods: qRT-PCR and in situ hybridization (ISH) was used to analyze LINC00339 expression in tumor tissues and cell lines. CCK8 and colony formation assays were used to analyze cell proliferation. Transwell assay was used to analyze cell migration and invasion. Xenograft experiment was used to test tumor growth in vivo.
Results: LINC00339 overexpression was correlated with an advanced stage, metastasis, and bad prognosis in HCC patients. Functional investigation showed that LINC00339 knockdown significantly suppressed HCC cell proliferation, migration, and invasion. Moreover, decreased LINC00339 expression inhibited HCC growth in vivo. Mechanistically, LINC00339 could interact with miR-1182 to promote SKA1 expression. We also demonstrated that SKA1 acted as an oncogene and SKA1 upregulation reversed the effect of LINC00339 silencing.
Conclusion: Our results illustrated that the LINC00339/miR-1182/SKA1 axis plays an essential role in HCC progression.
Keywords: LINC00339, miR-1182, SKA1, hepatocellular carcinoma, progression
Introduction
Hepatocellular carcinoma (HCC) is the most common cancer and remains a leading cause of cancer-associated death around the world.1 Nearly 600,000 cases are diagnosed as HCC patients each year.2 In spite of the advances in therapeutic approaches in the past decades, outcomes of HCC patients are unfavorable due to high rates of recurrence and metastasis.3 Hence, it is indispensable to investigate the pathogenesis of HCC development.
Long noncoding RNA (lncRNA) belongs to the noncoding RNA family and is over 200 nucleotides in length with poorly protein-coding potential.4 Several studies have shown that lncRNAs exert crucial functions in multiple tumors, including HCC.5,6 For example, lncPARP1 promotes HCC progression by upregulating PARP1.7 LncRNA SOX21-AS1 promotes HCC development by epigenetically inhibiting p21.8 LncRNA NEAT1 is an oncogene and promotes HCC proliferation.9 The aberrant expression of lncRNAs is usually related to HCC development and metastasis. For instance, lncWDR26 downregulation suppresses HCC proliferation and invasion.10 lncRNA HOXA-AS2 upregulation in HCC enhances tumor growth and induces epithelial–mesenchymal transition.11 Thus, because of the significant importance of lncRNAs, it is essential to explore the correlation between lncRNA and HCC progression.
LINC00339 is a poorly researched lncRNA. Previously, researchers have shown that LINC00339 promotes the development of glioma and laryngeal squamous cell carcinoma.12–14 However, the roles of LINC00339 in HCC are unclear. Our research indicated that LINC00339 was upregulated in HCC tissues. LINC00339 silencing impaired proliferation and metastasis through the miR-1182/SKA1 axis. Hence, we demonstrate from previously undefined signaling that the LINC00339/miR-1182/SKA1 axis modulates HCC progression.
Materials and methods
Human HCC tissues
We collected 62 HCC tissues from Wenzhou Central Hospital. All patients were not treated with chemotherapy or radiotherapy prior to collection. Samples were kept in liquid nitrogen. Our work was approved by the Ethics Committee of Wenzhou Central Hospital. Written informed consent was collected from every participant. Experiments involving human tissues were conducted in accordance with the Declaration of Helsinki.
Cell lines and transfection
HCC cell lines (HepG2, Hep3B, SMMC-7721, Huh7, and MHCC97H) and the normal hepatocyte cell line (LO2) were from the Institute of Biochemistry and Cell Biology (Shanghai, People’s Republic ofChina). The cell culture method has been reported before.15 Lentivirus was produced by GeneChemCo., Ltd. (Shanghai, People’s Republic of China). For lentivirus infection, HCC cells were plated in 6-well plates and 100 μL lentivirus (virus titer, 109 TU/ml) was added. Cells expressing green fluorescent protein were isolated and cultured for the following experiment. Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) was used for plasmid transfection.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNAs were extracted using TRIzol (Invitrogen). cDNA synthesis was carried out using a HiFi-MMLV cDNA Kit (CWBio, Co., Ltd.). Quantitative PCR was completed using the SYBR Premix ExTaq Reverse Transcription PCR kit (Takaka, Dalian, People's Republic of China). U6 was used for internal control. The 2−ΔΔCt method was used for expression calculation.
Cell proliferation detection
The CCK8 assay was used to detect cell viability.
Colony formation assay
A total of 500 cells were added to 6-well plates. Cells were cultured for 2 weeks. The colony was then fixed and stained, and the colony number was counted.
Migration and invasion assay
Transwell chambers were utilized to check migration and invasion. In brief, 2×105 cells were seeded into the upper chamber (precoated with Matrigel for invasion) with 200 μL serum-free medium. The bottom chamber was filled with 600 μL complete medium. After culture for 24 h, the migrated or invaded cells in the lower chamber were fixed and stained with crystal violet. The cell number was determined with an inverted microscope.
Animal studies
Four-week-old Balb/c nude female mice were obtained from Shanghai Laboratory Animal Company (Shanghai, People's Republic of China). Hep3B cells were injected subcutaneously into the right flank of nude mice. The tumor volumes were measured every week. After 5 weeks, the tumor tissues were removed and weighed. The animal experiments were approved by the Ethics Committee of Wenzhou Central Hospital. All animal operations were performed in accordance with the Animal Policy and Welfare Committee of our hospital.
Luciferase reporter assay
The potential binding sites were predicted using miRDB or TargetScan7. Then, the wild-type (wt) or mutant (mut) LINC00339 or SKA1 3-UTR sequences were constructed in pGL3 promoter vector (Promega Corporation, Fitchburg, WI, USA). For reporter assays, Hep3B cells were seeded in 24-well plates and cultured for 24 h. The luciferase reporter activity was then measured using the dual-luciferase reporter assay system (Promega). Renilla was used as the internal control for transfection efficiency.
RNA pulldown
The RNA pulldown assay was completed according to a previous report.16
Statistical analysis
Each result was presented as mean±SD. Significant differences were determined by Student’s t-test or one-way ANOVA using GraphPad Prism 6 software. p<0.05 was considered statistically significant.
Results
LINC00339 upregulation was observed
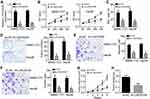
LINC00339 expression in HCC tissues was firstly analyzed. Through qRT-PCR, the LINC00339 level was found to be dramatically increased in HCC (Figure 1A). Then, we divided the samples into two subgroups based on TNM stages. qRT-PCR analysis revealed that LINC00339 levels were higher in the advanced stages of HCC tissues (Figure 1B). Besides, in situ hybridization assays were performed to examine the LINC00339 level in metastatic and nonmetastatic tissues. As shown, LINC00339 expression in metastatic tissues was elevated (Figure 1C), which was validated via PCR (Figure 1D). Next, LINC00339 expression was analyzed in tumor cell lines. Consistently, its levels were also increased in HCC cell lines (Figure 1E). Moreover, we found that HCC patients expressing a higher level of LINC00339 showed a relative lower survival rate (Figure 1F), indicating that LINC00339 may be an indicator for prognosis.
LINC00339 knockdown led to decreased HCC cell growth
SMMC-7721 and Hep3B cells were selected for the following experiments. We transduced these cells with shRNA against LINC00339 or negative controls. qRT-PCR results indicated that sh-LINC00339 transfection efficiently decreased the expression of LINC00339 (Figure 2A). LINC00339 knockdown gave rise to an impaired proliferation rate in SMMC-7721 and Hep3B cells (Figure 2B and C). LINC00339 depletion dramatically decreased the numbers of colonies (Figure 2D). Then, transwell assays were carried out. The results demonstrated that LINC00339 knockdown decreased cells of migration and invasion (Figure 2E and F). Thus, LINC00339 knockdown suppresses HCC proliferation and metastasis in vitro. To explore the roles of LINC00339 in vivo, we conducted a xenograft animal assay. LINC00339 silencing decreased the tumor volume and weight (Figure 2G and H). Thus, LINC00339 knockdown suppresses HCC growth in vivo.
LINC00339 targeted the miR-1182/SKA1pathway in HCC
Next, the mechanism of LINC00339 was explored. We identified miR-1182 as a possible target miRNA of LINC00339. Silencing of LINC00339 gave rise to upregulated miR-1182 (Figure 3A and B). The luciferase assay showed that the activity of wt-LINC00339 was suppressed by miR-1182 mimics in Hep3B cells (Figure 3C and D), supporting a direct interaction between miR-1182 and LINC00339. To further validate this, we performed the RNA pulldown assay using wt-miR-1182 and mut-miR-1182 probes. As shown, only wt-miR-1182 could precipitate LINC00339 in HCC cell lysates (Figure 3E and F). Besides, LINC00339 was negatively correlated with miR-1182 in HCC tissues (Figure 3G). Next, miR-1182 targets were further predicted. SKA1 was identified as the most potential candidate. We found that the luciferase activity of wt-SKA1 was inhibited after miR-1182 mimic transfection (Figure 3H and I), indicating that miR-1182 directly interacts with SKA1. Moreover, we found that miR-1182 mimics significantly suppressed SKA1 expression (Figure 3J). To further explore the correlation between SKA1 and LINC00339, we performed another luciferase reporter assay. LINC00339 depletion reduced the activity of wt-SKA1 reporter (Figure 3K). Besides, SKA1 expression was inhibited by LINC00339 knockdown in HCC cells (Figure 3L). Thus, our results demonstrated that LINC00339 is a ceRNA for miR-1182 to upregulate SKA1 expression in HCC.
SKA1 restoration rescued the effects of LINC00339 knockdown
Through analysis, we found that SKA1 was upregulated in HCC (Figure 4A and B). Furthermore, The Cancer Genome Atlas data also showed that SKA1 upregulation in HCC patients predicted a low survival rate (Figure 4C), suggesting a potential oncogenic role of SKA1. To validate whether SKA1 is the downstream effector of LINC00339, we performed rescue assays. We restored SKA1 expression (Figure 4D) and performed CCK8, EdU, and Transwell assays. The results showed that restoration of SKA1 significantly rescued the proliferation, migration, and invasion of LINC00339-depleted HCC cells (Figure 4E–H). Thus, our study demonstrated that the LINC00339/miR-1182/SKA1 axis modulates the progression of HCC.
Discussion
Our work illustrated essential roles of LINC00339 in HCC development. The LINC00339 level was found to be upregulated in HCC. An elevated level of LINC00339 was related to an advanced stage, metastasis, and poor prognosis. Through functional experiments, we demonstrated that LINC00339 suppressed proliferation, migration, and invasion of HCC cells in vitro and in vivo. Moreover, we found a novel mechanism. We reported that LINC00339 acts as the ceRNA for miR-1182 and promotes SKA1 expression. Taken together, our finding for the first time identified that the LINC00339/miR-1182/SKA1 axis contributes to HCC progression.
Accumulating research has confirmed the importance of lncRNAs in tumorigenesis.17,18 In HCC, several lncRNAs were identified as important regulators. For instance, SNHG16 overexpression suppresses HCC cell proliferation.19 Nevertheless, the functions of most lncRNAs in HCC are still unknown.20 Hence, defining the functions and mechanism is crucial for the development of lncRNA-related therapeutic approaches. LINC00339 was the first lncRNA reported to regulate glioma.12 Then, Yuan et al21 reported that LINC00339 promotes the development and progression of nonsmall cell lung cancer via the miR-145/FOXM1 axis. Recently, several researchers have demonstrated that LINC00339 promotes proliferation, migration, and invasion of laryngeal squamous cell carcinoma and breast cancer.13,14 Its role in HCC remains limitedly studied. We provided evidence that LINC00339 promotes HCC tumorigenesis and metastasis through the miR-1182/SKA1 pathway. We demonstrated that LINC00339 promotes HCC proliferation. Through the Transwell assay, we indicated that LINC00339 knockdown suppressed migration and invasion. Moreover, a xenograft mouse model suggested that LINC00339 knockdown inhibited growth of HCC in vivo. Thus, our work for the first time demonstrated the oncogenic roles of LINC00339 in HCC.
Increasing studies have shown that lncRNAs could work as ceRNAs to regulate biological processes.22 For instance, A1BG-AS1 interacts with miR-216a-5p to regulate HCC development.23 Previous studies have reported that LINC00339 is the sponge for miR-377-3p, miR-145, and miR-539-5p.12–14 This work for the first time identified LINC00339 as the sponge for miR-1182. To date, knowledge about miR-1182 is poor. Zhang et al24 indicated that miR-1182 inhibits gastric cancer growth and invasion. Zhou et al25 reported that miR-1182 represses bladder cancer growth. Besides, Hou et al26 proved that miR-1182 attenuates ovarian cancer proliferation and invasion. How miR-1182 regulates HCC, however, is unknown. We demonstrated that miR-1182 was directly sponged by LINC00339 in HCC. Moreover, we demonstrated that LINC00339 suppressed the levels of miR-1182 in HCC cells and indicated that miR-1182 was a potential tumor suppressor for HCC.
Afterward, SKA1 was validated to be the downstream target of the LINC00339/miR-1182 axis. Previous studies have shown that SKA1 downregulation suppresses proliferation of bladder cancer and adenoid cystic carcinoma.27,28 A report also indicated that SKA1 overexpression predicts poor prognosis in HCC.29 Nevertheless, how SKA1 modulates HCC remains undefined. The present work showed that SKA1 expression was modulated by the LINC00339/miR-1182 axis. Then, we also showed that SKA1 was upregulated in HCC tissues and indicated poor prognosis. Finally, we performed a rescue assay. We found that restoration of SKA1 rescued the abilities of proliferation, migration, and invasion. Thus, our study for the first time demonstrated that SKA1 regulates the malignant behaviors of HCC cells.
In summary, we identified that LINC00339 promotes HCC progression by targeting the miR-1182/SKA1 axis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zamor PJ, deLemos AS, Russo MW. Viral hepatitis and hepatocellular carcinoma: etiology and management. J Gastrointest Oncol. 2017;8(2):229–242. doi:10.21037/jgo.2017.03.14
2. Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi:10.1245/s10434-011-1581-y
3. Wang P, Ouyang L, Zheng L, Wang Z. Identifying hepatocellular carcinoma-related genes and pathways by system biology analysis. Ir J Med Sci. 2015;184(2):357–364. doi:10.1007/s11845-014-1119-y
4. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
5. Yu FJ, Zheng JJ, Dong PH, Fan XM. Long non-coding RNAs and hepatocellular carcinoma. Mol Clin Oncol. 2015;3(1):13–17. doi:10.3892/mco.2014.429
6. Zhu P, Wang Y, Wu J, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi:10.1038/ncomms13608
7. Qi H, Lu Y, Lv J, et al. The long noncoding RNA lncPARP1 contributes to progression of hepatocellular carcinoma through up-regulation of PARP1. Biosci Rep. 2018;38(3):BSR20180703. doi:10.1042/BSR20180703
8. Wei C, Wang H, Xu F, Liu Z, Jiang R. LncRNA SOX21-AS1 is associated with progression of hepatocellular carcinoma and predicts prognosis through epigenetically silencing p21. Biomed Pharmacother. 2018;104:137–144. doi:10.1016/j.biopha.2018.05.010
9. Liu X, Liang Y, Song R, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018;17(1):90. doi:10.1186/s12943-018-0838-5
10. Chen B. A novel long noncoding RNA lncWDR26 suppresses the growth and metastasis of hepatocellular carcinoma cells through interaction with SIX3. Am J Cancer Res. 2018;8(4):688–698.
11. Zhang Y, Xu J, Zhang S, et al. HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition via the miR-520c-3p/GPC3 axis in hepatocellular carcinoma. Cell Physiol Biochem. 2018;50(6):2124–2138. doi:10.1159/000495056
12. Guo J, Cai H, Liu X, et al. Long non-coding RNA LINC00339 stimulates glioma vasculogenic mimicry formation by regulating the miR-539-5p/TWIST1/MMPs axis. Mol Ther Nucleic Acids. 2018;10:170–186. doi:10.1016/j.omtn.2017.11.011
13. Liu S, Duan W. Long noncoding RNA LINC00339 promotes laryngeal squamous cell carcinoma cell proliferation and invasion via sponging miR-145. J Cell Biochem. 2018;119:9694–9706. doi:10.1002/jcb.27284
14. Wang X, Chen T, Zhang Y, et al. Long noncoding RNA Linc00339 promotes triple-negative breast cancer progression through miR-377-3p/HOXC6 signaling pathway. J Cell Physiol. 2019;234(8):13303–13317. doi:10.1002/jcp.28007
15. Li Y, Liu G, Li X, Dong H, Xiao W, Lu S. Long non-coding RNA SBF2-AS1 promotes hepatocellular carcinoma progression through regulation of miR-140-5p-TGFBR1 pathway. Biochem Biophys Res Commun. 2018;503(4):2826–2832. doi:10.1016/j.bbrc.2018.08.047
16. Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233(5):4044–4055. doi:10.1002/jcp.26072
17. Gao J, Zeng K, Liu Y, Gao L, Liu L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 2019;12:169–179. doi:10.2147/OTT.S184078
18. Wang Y, Zhang G, Han J. HIF1A-AS2 predicts poor prognosis and regulates cell migration and invasion in triple-negative breast cancer. J Cell Biochem. 2019;120(6):10513–10518. doi:10.1002/jcb.28337
19. Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855–8863. doi:10.2147/OTT.S182005
20. Niu ZS, Niu XJ, Wang WH. Long non-coding RNAs in hepatocellular carcinoma: potential roles and clinical implications. World J Gastroenterol. 2017;23(32):5860–5874. doi:10.3748/wjg.v23.i32.5860
21. Yuan Y, Haiying G, Zhuo L, Ying L, Xin H. Long non-coding RNA LINC00339 facilitates the tumorigenesis of non-small cell lung cancer by sponging miR-145 through targeting FOXM1. Biomed Pharmacother. 2018;105:707–713. doi:10.1016/j.biopha.2018.06.022
22. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
23. Bai J, Yao B, Wang L, et al. lncRNA A1BG-AS1 suppresses proliferation and invasion of hepatocellular carcinoma cells by targeting miR-216a-5p. J Cell Biochem. 2019;120(6):10310–10322. doi:10.1002/jcb.28315
24. Zhang D, Xiao YF, Zhang JW, et al. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015;360(2):151–159. doi:10.1016/j.canlet.2015.01.044
25. Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun. 2016;470(2):445–452. doi:10.1016/j.bbrc.2016.01.014
26. Hou XS, Han CQ, Zhang W. MiR-1182 inhibited metastasis and proliferation of ovarian cancer by targeting hTERT. Eur Rev Med Pharmacol Sci. 2018;22(6):1622–1628. doi:10.26355/eurrev_201803_14569
27. Tian F, Xing X, Xu F, et al. Downregulation of SKA1 gene expression inhibits cell growth in human bladder cancer. Cancer Biother Radiopharm. 2015;30(7):271–277. doi:10.1089/cbr.2014.1715
28. Zhao LJ, Yang HL, Li KY, et al. Knockdown of SKA1 gene inhibits cell proliferation and metastasis in human adenoid cystic carcinoma. Biomed Pharmacother. 2017;90:8–14. doi:10.1016/j.biopha.2017.03.029
29. Chen YB, Zhao JJ, Jiao ZH, et al. SKA1 overexpression is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer. 2018;18:1240.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.