Back to Journals » Cancer Management and Research » Volume 13
LINC00265/miR-4500 Axis Accelerates Acute Lymphoblastic Leukemia Progression by Enhancing STAT3 Signals
Authors Zhao D, Xing Q, Song H, Zhao Y, Guo G
Received 29 July 2020
Accepted for publication 1 October 2020
Published 28 October 2021 Volume 2021:13 Pages 8147—8156
DOI https://doi.org/10.2147/CMAR.S274590
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Donglu Zhao, Qi Xing, Hang Song, Yan Zhao, Guiying Guo
No. 4 Ward of Hematology Department, Institute of Hematology and Oncology, Harbin First Hospital, Harbin 150010, Heilongjiang Province, People’s Republic of China
Correspondence: Donglu Zhao
No. 4 Ward of Hematology Department, Institute of Hematology and Oncology, Harbin First Hospital, No. 149 Diduan Street, Daoli District, Harbin 150010, Heilongjiang Province, People’s Republic of China
Tel +86-13936254716
Email [email protected]
Background: Long noncoding RNA LINC00265 or miR-4500 is involved in the pathogenesis of many cancers. However, their functions in acute lymphoblastic leukemia (ALL) remain unknown. In this study, we investigated how LINC00265 and miR-4500 regulate the malignant characteristics of ALL.
Methods: Real-time PCR was used in examining the expression of LINC00265 in ALL cell lines and blood of patients with ALL. Cell proliferation, cell migration, and xenograft tumor assays were performed to verify the function of LINC00265 subjected to overexpressing and silencing experiments. The ceRNA mechanism with LINC00265/miR-4500/STAT3 was investigated through luciferase and RNA pull-down assays. Finally, the function of the LINC00265/miR-4500/STAT3 axis subjected to overexpressing and silencing assays was determined through cell proliferation, cell migration, and xenograft tumor assays.
Results: LINC00265 was highly expressed in ALL cell lines and blood of patients with ALL and facilitated the proliferation, migration, invasion, and growth of xenograft tumors of ALL cells. The silencing of LINC00265 expression with LINC00265 siRNA significantly inhibited the malignancy of the ALL cells. RNA pull-down and luciferase assays demonstrated that LINC00265 competitively targeted miR-4500 and enhanced STAT3 expression. Furthermore, miR-4500 inhibitors or overexpressed LINC00265 up-regulated STAT3 expression, and miR-4500 mimics or STAT3 shRNAs eliminated the LINC00265-induced malignancy of the ALL cells.
Conclusion: Mechanistically, LncRNA LINC00265 can competitively interact with miR-4500 and thereby up-regulates STAT3 signaling and enhances the malignancy of tumors.
Keywords: acute lymphoblastic leukemia, LINC00265, miR-4500, cell proliferation
Introduction
Acute lymphoblastic leukemia (ALL) is a hematologic malignancy driven by the proliferation and accumulation of a group of lymphoid neoplasms that morphologically and immunophenotypically resemble B-lineage and T-lineage precursor cells.1,2 The incidence of ALL is 1.7 cases per 100,000 individuals per year in the USA.3,4 Approximately 70% of leukemia cases in children are ALL,5 and ALL accounts for 20% of leukemia cases in adults.3,6 The etiology and pathogenesis of ALL are not fully understood. Thus, elucidating the molecular mechanisms underlying the pathogenesis of the disease is important to the formulation of novel therapeutic strategies for ALL.
Widespread evidence demonstrates that long noncoding RNAs (lncRNAs) play important roles in diverse biological and pathological processes.7 Many lncRNAs have been identified as the regulators of four main types of leukemia, namely, acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia, and chronic myeloid leukemia.8 For example, the lncRNAs MEG3, Xist, HOTAIR, and LINC002659,10 are involved in the pathological processes of AML, and CDKN2B-AS, LUNAR1, and LINC00958 are involved in the pathological processes of ALL.8 LINC00265 may regulate the proliferation, migration, and invasion function of AML cells via PI3K/AKT signaling10 and can mediate autophagy suppressing the apoptosis of acute myeloid leukemia cells by miR-485-5p/IRF2 axis.9 However, the regulation of ALL by LINC00265 has not been elucidated.
Another kind of small noncoding RNAs, microRNAs (miRNAs), also play an important role in numerous pathogenesis of cancers.11 miR-708-5p, miR-195-5p, miR-450b-5p, and miR-29c-5p act as oncogenes or tumor suppressors in T- or B-cell ALL progression by regulating cells proliferation, metastasis, and apoptosis.12 miRNA-326,13 miRNA-9,14 miR-153-3p,15 miRNA-452,16 and miRNA-200c13 are involved in ALL processes and can be used as targets for the diagnosis, prognosis, and treatment of ALL. miRNA-4500 has been recently found to be a cancer suppressor that can inhibit cell proliferation and migration in bladder cancer,17 papillary thyroid cancer,18 and breast cancer,19 but its function in ALL remains unknown. Aberrant STAT3 activation promotes tumorigenesis through oncogenic gene expression in various cancers, enhancing tumor malignancy.20 It evolves as an important target for potential therapeutic strategies for cancers.
In the study, we explore novel lncRNAs to modulate ALL pathogenesis through sponging miRNAs. We first found that LINC00265 expression increased in ALL cell lines, especially in the blood of patients with ALL, and facilitated the proliferation, migration, invasion, and growth of the xenograft tumors of ALL cells. Furthermore, it can competitively interact with miR-4500, up-regulate downstream signaling STAT3, and enhance the malignancy of tumors. Hence, it is a potential target molecule for treating and diagnosing ALL.
Materials and Methods
Blood Samples of Patients
Fifty patients with acute lymphoblastic leukemia were selected from the Department of Hematology at Harbin First Hospital from June 2018 to August 2020. Diagnosis was confirmed by bone marrow cell morphology and immunophenotypic examination. All the enrolled patients (30 males and 20 females) were newly treated leukemia patients. Of these patients, 12 had T-cell lymphocytic leukemia (T-ALL) cases, and 38 had B-cell lymphocytic leukemia. The average age was 42.45±6.5 years. The inclusion criteria were as follows: diagnostic criteria for ALL are met and clinical manifestations are anemia, fever, infection, and hemorrhage. The exclusion criteria were as follows: pregnancy; mental diseases, unable to cooperate with researchers; and chronic myelogenous leukemia with acute change. In the control group, 50 volunteers were selected from a healthy outpatient physical examination group. None of the participants had recent infection, bleeding, or immunosuppressive medication, and differences in age and sex between the acute lymphoblastic leukemia group and the normal group were nonsignificant. T cells were isolated with an EasySep human T cell isolation kit (STEMCELL Technologies, Vancouver, BC, Canada), and the samples were snap-frozen in liquid nitrogen and stored at −80°C. All the studies have been approved by the Ethics Committee of Harbin First Hospital, and informed consents were signed by participants.
Cell Culture
Human T-cell acute lymphoblastic leukemia cell lines, Molt-3, Molt-4, CCRF-CEM, TALL104, human acute lymphoblastic leukemia cell lines, and RS411 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in RPMI-1640 media supplemented with 10% FBS and 1% penicillin/streptomycin (complete media) and incubated at 37°C and 5% CO2. SUP-B15 human B-cell acute lymphoblastic leukemia cell lines were cultured in Iscove’s Modified Dulbecco’s Medium with 4 mM L-glutamine adjusted to contain 1.5 g/l sodium bicarbonate, supplemented with 0.05 mM 2-mercaptoethanol and 20% FBS, and incubated at 37°C and 5% CO2.
Cell Transfection
LINC00265 siRNA and negative control, miR-4500 mimics and inhibitor, STAT3 siRNA and negative control were purchased from Ribobio (Guangzhou, China). LINC00265 or STAT3 wild-type (WT) and mutant-type (MT) plasmids and corresponding blank plasmids were prepared in our laboratory. Molt-3 and SUP-B15 cells were seeded in 12-wells plates and cultured for 12 h before transfection. For transient transfection, LINC00265 siRNA, miR-4500 mimics, STAT3 siRNA, and recombinant plasmid were transfected using Lipofectamine 2000 (Invitrogen). Samples were collected after 24 h for the quantification of target gene expression.
Real-Time PCR
Tissue samples or cells were homogenized in Trizol, and total RNA extraction and reverse transcription were performed according to the manufacturer’s protocol. For the PCR amplification of the cDNA fragment coding target genes, sense and antisense primer sequences for LINC00265, miR-4500, STAT3, U6, and β-actin were as follows: β-actin, F: 5ʹ-GATCTTGATCTTCATGGTGCTAG-3ʹ, R: 5ʹ-TTGTAACCACCTGGGACGATATGG-3ʹ. U6, F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. RT-qPCR was conducted using the miScript SYBR® Green PCR Kit (Thermo, USA). Relative expression levels were calculated using 2−ΔΔCt method.
Luciferase Assays
The complementary and mutant sequences of LINC00265 and STAT3 with an miR-4500 binding site were constructed into dual-luciferase reporter vectors. HEK 293T cells were co-transfected with miR-4500 or control mimics, firefly luciferase reporter vector containing WT or mutant (Mut) sequences of LINC00265 and STAT3 using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Renilla and firefly luciferase activities were examined 24 h post-transfection with a Dual-Luciferase® reporter assay system.
Cell Counting Kit-8 Assay
MOLT3 or SUP-B15 cells were seeded in a 96-well plate (3000/well). Cell proliferation was examined using Cell Counting Kit-8 (CCK-8) according to the manufacturer’s instructions. Absorbance at 450 nm 48 h after seeding was measured with a microplate reader (Bio-Rad, CA).
TUNEL Assays
The TUNEL assay kit used was purchased from Abcam corporation. Cells were treated and fixed cells in 4% PFA, then permeabilized for 2 min with 0.1% Triton X-100. The cells were incubated with a TUNEL reaction mixture in the dark for 1 h, then with Hoechst 33,342 for 15 min. The cells were mounted with ProLong Gold antifade overnight and imaged with a fluorescence microscope.
Cell Migration Assays
MOLT3 or SUP-B15 cells were transfected with LINC00265 and control siRNA; LINC00265 recombinant and blank plasmid; LINC00265 and miR-4500 mimics; or LINC00265 and sh-STAT3 in 12-well plates (10,000 cells/well) for 24 h. Then, 5000 cells per well were seeded in the cell culture dish (Corning, USA). After 24 h of incubation, the inserts were rinsed, fixed in 2% paraformaldehyde for 10 min, and stained with crystal violet (Beyotime, Jiangsu, China). Migrated cells were counted under a microscope (ECLIPSE Ti, Nikon, Japan).
RNA Pull-Down Assay
The LINC00265 and miR-4500 were biotin-labeled into biotin-miR-4500 and biotin-LINC00265, and then incubated with cell lysates before streptavidin-coated magnetic beads (Life Technologies) were added. Pull-down assay was performed in biotin-coupled RNA complex, and the quantities of LINC00265, STAT3, and miR-4500 were determined through qPCR.
Xenograft Tumor
Ten female nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) and inoculated subcutaneously (s.c.) injection with 5×107 mL−1 (100 μL) Lenti-NC- and Lenti-LINC00265-treated cells successively on the right leg. Tumor volumes were measured from day 10 to day 24, and the mice received an intraperitoneal injection of 0.5% pentobarbital sodium and suffocated with carbon dioxide. Subsequently, the xenograft tumors were removed surgically and weighted. All the animal experiments were approved by the animal ethics committee of Harbin First Hospital and performed in the Harbin First Hospital. All experiments followed the guideline of the ethical review of animal welfare (GB T 35,892–2018).
Statistical Analysis
Each experiment was performed in triplicate, and data were analyzed with GraphPad Prism 8.0 software. Two-tailed test, unpaired Student’s t-test, or one-way ANOVA were performed. Differences with P values of less than 0.05 were considered statistically significant.
Results
LINC00265 Was Up-Regulated in ALL Patients and Cell Lines
To evaluate the expression of LINC00265 in ALL patients and cell lines, we tested the expression of LINC00265 in the blood and ALL cell lines. We first collected 35 paired blood samples from patients with ALL and healthy people and then examined the expression of LINC00265 through quantitative PCR (qPCR). Real-time PCR data showed that LINC00265 was remarkably increased in the blood of patients with ALL compared with that in the blood of healthy people (Figure 1A). Furthermore, we found that LINC00265 was highly expressed in the ALL cell lines compared with that in peripheral blood mononuclear cells (PBMCs) and bone marrow mononuclear cells (BMMCs) from healthy people (Figure 1B). These results suggested that LINC00265 is highly expressed in the blood of the patients with ALL and ALL cell lines.
Linc00265 Facilitates the Proliferation, Migration, and Invasion of ALL Cell Lines MOLT3 and SUP-B15
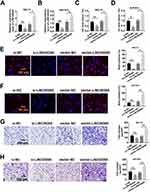
To uncover the function of LINC00265 in ALL progression, we treated MOLT3 or SUP-B15 cells with LINC00265 siRNA. We found that the treatment significantly down-regulated the expression of LINC00265 (Figure 2A and B) and cell viability remarkably decreased (Figure 2C and D). After the expression of LINC00265 was enhanced with LINC00265-overexpressing plasmid in the MOLT3 or SUP-B15 cells (Figure 2A and B), cell viability considerably increased (Figure 2C and D). Cell proliferation capacity was examined using the BrdU assay. The positive cells of BrdU in MOLT3 or SUP-B15 cells increased in the LINC00265 siRNA-treated group compared with those in the siRNA negative control group (Figure 2E and F). The number of positive cells of BrdU in MOLT3 or SUP-B15 cells treated with LINC00265-overexpressing plasmid showed a higher increase than that of the same cells treated with siRNA negative control (Figure 2E and F). In addition, we determined the effect of LINC00265 on cell invasion through Transwell assay. We found that the number of MOLT3 or SUP-B15 cells in the LINC00265 siRNA-treated group was lower than that in the siRNA negative control group (Figure 2G and H). By contrast, the number of MOLT3 or SUP-B15 cells in the LINC00265 recombinant plasmid-treated group exhibited an increase compared with that in the blank plasmid-treated group (Figure 2G and H). These data indicated that LINC00265 facilitates the proliferation, migration, and invasion of ALL cells.
LINC00265 Promotes the Growth of MOLT3 Xenograft Tumor
To investigate the role of LINC00265 in tumor growth, we packaged LINC00265 overexpression lentivirus to infect MOLT3 cells and injected 1×106 infected cells subcutaneously into 8-week-old nude mice. The MOLT3 xenograft tumor treated with lenti-LINC00265 was remarkably large compared with that treated with lenti-negative control (Figure 3A). We then measured the volumes of the xenograft tumors from day 10 to day 24 after injection and found that tumor volume in the lenti-LINC00265 group was substantially larger than that in the lenti-negative control group (Figure 3B). In addition, we found that the tumor mass of the lenti-LINC00265 group was considerably higher than that of the lenti-negative control group (Figure 3C). These results suggested that LINC00265 can promote the growth of MOLT3 xenograft tumors.
LINC00265 Interacts with miR-4500
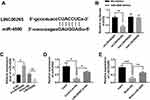
As is well known, competing endogenous RNAs (ceRNAs) can regulate target genes by interacting with lncRNAs and miRNAs. We performed alignment assays to determine whether LINC00265 has a function of sponging miRNAs similar to ceRNAs. The results showed that miR-4500 targeted LINC00265 (Figure 4A). To further confirm the interaction between LINC00265 and miR-4500, we constructed a luciferase reporter with LINC00265 WT and Mut sequences for miR-4500. The co-transfection of LINC00265-WT and miR-4500 in HEK 293T cells suppressed luciferase activity, whereas the co-transfection of AK087124-Mut and miR-4500 abolished the repression of luciferase activity (Figure 4B). To further determine the expression of miR-4500 in cells treated with LINC00265 siRNA or recombinant plasmid, we found that LINC00265 siRNA can increase miR-4500 expression level, whereas LINC00265 overexpression in the plasmid-treated groups remarkably decreased the miR-4500 expression level (Figure 4C). We also performed the RNA pull-down assay to detect the interaction between LINC00265 and miR-4500. As shown in Figure 4D and E, the LINC00265 and miR-4500 biotin probe remarkably enriched more counterparts than their negative control probes. These findings demonstrated that LINC00265 is involved in ALL progression particularly by targeting miR-4500 and negatively interacting with it.
miR-4500 Targets STAT3 to Silence Its Expression
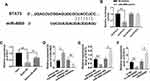
To identify the target gene of miR-4500, we performed bioinformatic analysis via alignment and found that STAT3 is a molecular target for miR-4500 because STAT3 3′UTR is complementary to the sequences of miR-4500 (Figure 5A). Furthermore, STAT3 WT and Mut 3ʹUTR were constructed into a dual-luciferase reporter system and used in determining whether miR-4500 directly targets STAT3 3′UTR. We found that the co-transfection of miR-4500 and STAT3 3′UTR WT in 293T cells significantly down-regulated luciferase activity compared with miRNA negative control. The co-transfection of STAT3 3ʹUTR Mut and miR-4500 was unable to decrease luciferase activity (Figure 5B). The interaction between miR-4500 and STAT3 was characterized using RNA pull-down assays. As shown in Figure 5C, the miR-4500 biotin probe substantially enriched more STAT3 compared with the negative control probe. We then treated the cells with miR-4500 mimics and inhibitors. The qPCR results showed that miR-4500 mimics were effectively transfected into the cells and significantly silenced the expression of STAT3 and miR-4500 inhibitors effectively inhibited miR-4500 mimics and considerably increased the expression of STAT3 (Figure 5D and E). Moreover, LINC00265 siRNA obviously inhibited the expression of STAT3, and overexpressed LINC00265 substantially promoted the expression of STAT3 (Figure 5F). These data suggested that miR-4500 participates in the regulation of ALL progress by targeting STAT3.
Overexpressed miR-4500 or Knockdown STAT3 Could Reverse ALL Progression by LINC00265
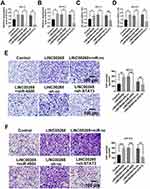
To understand the biological function of the LINC00265/miR-4500/STAT3 axis in ALL progression, we first determined the relationship among LINC00265, miR-4500, and STAT3. LINC00265 can enhance the expression of STAT3, whereas miR-4500 mimics and LINC00265 can significantly down-regulate the expression of STAT3 in MOLT3 or SUP-B15 cells compared with miR-4500 control and LINC00265 (Figure 6A and B). When MOLT3 or SUP-B15 cells were treated with STAT3 siRNA and LINC00265 STAT3 expression remarkably decreased compared with that in STAT3 siRNA negative control or LINC00265-treated group (Figure 6A and B). CKK-8 assays revealed that cell survival capacity in MOLT3 or SUP-B15 cells treated with miR-4500 mimics and LINC00265 were down-regulated compared with that in miR-4500 control and LINC00265-treated groups (Figure 6C and D). When MOLT3 or SUP-B15 cells were treated with STAT3 siRNA and LINC00265, cell survival capacity decreased compared with that in the STAT3 shRNA negative control and LINC00265-treated groups (Figure 6C and D). In addition, we examined the effect of LINC00265/miR-4500/STAT3 axis on cell invasion through Transwell assay. The results showed that compared with LINC00265 + miR-nc-treated group, the LINC00265 + miR-4500-treated group showed a decrease in the number of invasion cells (Figure 6E and F). When MOLT3 or SUP-B15 cells were treated with STAT3 shRNA and LINC00265, invasion cell number decreased compared with that in the STAT3 siRNA negative control and LINC00265-treated groups (Figure 6E and F). These findings indicated that LINC00265 facilitates the proliferation, migration, and invasion of ALL cells.
Discussion
Our study first demonstrated that LINC00265 can act as a competing endogenous RNA on miR-4500 and enhance the malignant characteristics of ALL, such as cancer cell proliferation, migration, and invasion and xenograft tumor growth, by increasing STAT3 expression. LINC00265 was highly expressed in the blood of patients with ALL and ALL cell lines, and the silencing of LINC00265 expression with LINC00265 siRNA significantly inhibited the malignancy of tumor cells. The miR-4500 mimics silenced the expression of STAT3 and promoted the progression of ALL.
lncRNAs are the most widespread and important noncoding RNAs that have been extensively investigated in various cancers,21 including ALL.22 LINC00265 and SPRY4-IT1 are highly expressed as oncogenes, whereas TRIM52-AS1 and BX357664 have low expression levels as tumor suppressors involved in tumorigenesis.18,23,24 The lncRNA uc.112 is highly expressed in ALL as an oncogene,25 and TCL6 as a tumor suppressor in ALL can significantly increase disease-free survival in “TCL6 high” patients compared with “TCL6 low” patients.26 As far as we know, LINC00265 has not been explored in ALL. Our study first demonstrated that LINC00265 is highly expressed as an oncogene in ALL. When artificially overexpressed in renal cancer cell lines, LINC00265 enhances the proliferation, migration, invasion, and growth of xenograft tumors of ALL cells.
miRNAs have been extensively investigated in various cancers.11 MiRNA-25 as an oncogene is highly expressed,27 and miRNA-4500 as a tumor suppressor involved in tumorigenesis has a low expression level.19 miR-4500 can inhibit the STAT3/CCR7 pathway and thereby suppresses cell proliferation and migration in bladder cancer.17 miR-4500 can directly target and repress PLXNC1 and inhibit the malignant transformation of papillary thyroid cancer cells.18 In addition, miR-4500 can decrease RRM2 level and inhibit the MAPK signaling pathway, thereby attenuating breast cancer cell proliferation, invasion, and migration and promote breast cancer cell apoptosis.19 Currently, the function of miR-4500 is unclear. Our investigation first found that miR-4500 can target STAT3 3′UTR to inhibit the malignancy of ALL.
As a ceRNA, LINC00265 can competitively interact with other miRNAs and regulate tumorigenesis. LINC00265 promotes the progression of colorectal cancer by regulating the miR-216b-5p/TRIM44 Axis28 and is overexpressed in AML cells. It suppresses the proliferation, migration, and invasion of AML cell lines by activating PI3K/AKT signaling.10 Furthermore, LINC00265 functions as a ceRNA for miR-485-5p and facilitates IRF2 expression, thereby suppressing apoptosis in acute myeloid leukemia cells.9 We demonstrated that LINC00265 can competitively sponge miR-4500 and accelerate ALL progression by enhancing STAT3 signals. In addition, LINC00511 exacerbates T-ALL progression via the miR-195-5p/LRRK1 axis, provides a potential therapeutic target for patients with T-ALL.29 LINC00221 regulates ATP2A2 expression by sponging miR-152-3p, exhibiting anti-proliferation and pro-apoptosis effects in ALL cells.30
In the study, we found that miR-4500 inhibitors or overexpressed LINC00265 can up-regulate STAT3 expression, whereas miR-4500 mimics or STAT3 shRNAs eliminate the LINC00265-induced malignancy of ALL cells, specifically suppressing ALL cell proliferation and invasion. These results are consistent with the function of STAT3 in breast cancer31 and head and neck cancer,32 although they should be further examined in clinical samples.
The limitations of this study include the absence of further discussion of the effects of the LINC00265/miR-4500/STAT3 axis on downstream signaling pathways, and whether LINC00265 can be used as a marker for clinical diagnosis needs to be further studied. A large sample size should be used.
Conclusions
We demonstrated for the first time that LINC00265 can competitively interact with miR-4500 and facilitates the proliferation, invasion, and growth of the xenograft tumors of ALL cells by enhancing STAT3 signaling. Thus, they are potential diagnostic biomarkers and therapeutic targets for ALL.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Gavralidis A, Brunner AM. Novel therapies in the treatment of adult acute lymphoblastic leukemia. Curr Hematol Malig Rep. 2020. doi:10.1007/s11899-020-00591-4
2. Jabbour E, O’Brien S, Konopleva M, Kantarjian H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer. 2015;121(15):2517–2528. doi:10.1002/cncr.29383
3. Phelan KW, Advani AS. Novel therapies in acute lymphoblastic leukemia. Curr Hematol Malig Rep. 2018;13(4):289–299. doi:10.1007/s11899-018-0457-7
4. Short NJ, Kantarjian H, Jabbour E, Ravandi F. Novel therapies for older adults with acute lymphoblastic leukemia. Curr Hematol Malig Rep. 2018;119(2):91–99. doi:10.1002/jcb.26800
5. Hunger SP, Mullighan CG, Longo DL. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. doi:10.1056/NEJMra1400972
6. Portell CA, Advani AS. Novel targeted therapies in acute lymphoblastic leukemia. Leuk Lymphoma. 2014;55(4):737–748. doi:10.3109/10428194.2013.823493
7. Ng M, Heckl D, Klusmann J-H. The regulatory roles of long noncoding RNAs in acute myeloid leukemia. Front Oncol. 2019;9:570. doi:10.1038/s41408-019-0258-9
8. Liu Y, Sun P, Zhao Y, Liu B. The role of Long Non-Coding RNAs (lncRNAs) and downstream signaling pathways in leukemia progression. Hematol Oncol. 2020. doi:10.1002/hon.2776
9. Zhang F, Li Q, Zhu K, et al. LncRNA LINC00265/miR-485-5p/IRF2-mediated autophagy suppresses apoptosis in acute myeloid leukemia cells. Am J Transl Res. 2020;12:2451–2462.
10. Ma L, Kuai WX, Sun XZ, Lu XC, Yuan YF. Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:7867–7876. doi:10.26355/eurrev_201811_16412
11. Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. doi:10.1093/carcin/bgx026
12. Almeida RS, Costa e Silva M, Coutinho LL, et al. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol Oncol. 2019;37(1):103–112. doi:10.1002/hon.2567
13. Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119(7):6024–6032. doi:10.1002/jcb.26800
14. Zang Y, Yu R, Bai Y, Chen X. MicroRNA-9 suppresses cancer proliferation and cell cycle progression in acute lymphoblastic leukemia with inverse association of neuropilin-1. J Cell Biochem. 2018;119(8):6604–6613. doi:10.1002/jcb.26799
15. Jiang J, Liu Y, Zhao Y, Tian F, Wang G. miR-153-3p suppresses inhibitor of growth protein 2 expression to function as tumor suppressor in acute lymphoblastic leukemia. Technol Cancer Res Treat. 2019;18:1533033819852990. doi:10.1177/1533033819852990
16. Wang H, Guo Q, Zhu G, et al. microRNA-452 exerts growth-suppressive activity against T-cell acute lymphoblastic leukemia. J Investig Med. 2018;66(4):773–779. doi:10.1136/jim-2017-000591
17. Peng W, Dong N, Wu S, et al. miR-4500 suppresses cell proliferation and migration in bladder cancer via inhibition of STAT3/CCR7 pathway. J Cell Biochem. 2019. doi:10.1002/jcb.29558
18. Ge H, Yan Y, Yue C, Liang C, Wu J. Long noncoding RNA LINC00265 targets EGFR and promotes deterioration of colorectal cancer: a Comprehensive Study based on data mining and in vitro validation. Onco Targets Ther. 2019;12:10681–10692. doi:10.2147/OTT.S227482
19. Li S, Mai H, Zhu Y, et al. MicroRNA-4500 inhibits migration, invasion, and angiogenesis of breast cancer cells via RRM2-dependent MAPK signaling pathway. Mol Ther Nucleic Acids. 2020;21:278–289. doi:10.1016/j.omtn.2020.04.018
20. Lee H, Jeong AJ, Ye S-K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019;52(7):415–423. doi:10.5483/BMBRep.2019.52.7.152
21. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi:10.1016/j.gpb.2015.09.006
22. James AR, Schroeder MP, Neumann M, et al. Long non-coding RNAs defining major subtypes of B cell precursor acute lymphoblastic leukemia. J Hematol Oncol. 2019;12(1):8. doi:10.1186/s13045-018-0692-3
23. Liu X, Hao Y, Yu W, et al. Long non-coding RNA emergence during renal cell carcinoma tumorigenesis. Cell Physiol Biochem. 2018;47(2):735–746. doi:10.1159/000490026
24. Li M, Wang Y, Cheng L, et al. Long non-coding RNAs in renal cell carcinoma: a systematic review and clinical implications. Oncotarget. 2017;8(29):48424–48435. doi:10.18632/oncotarget.17053
25. Das Chagas PF, de Sousa GR, Kodama MH, et al. Ultraconserved long non-coding RNA uc.112 is highly expressed in childhood T versus B-cell acute lymphoblastic leukemia. Hematol Transfus Cell Ther. 2020. doi:10.1016/j.htct.2019.12.003
26. Cuadros M, Andrades Á, Coira IF, et al. Expression of the long non-coding RNA TCL6 is associated with clinical outcome in pediatric B-cell acute lymphoblastic leukemia. Blood Cancer J. 2019;9(12):93. doi:10.1038/s41408-019-0258-9
27. Frixa T, Donzelli S, Blandino G. Oncogenic MicroRNAs: key players in malignant transformation. Cancers. 2015;7(4):2466–2485. doi:10.3390/cancers7040904
28. Sun S, Li W, Ma X, Luan H. Long noncoding RNA LINC00265 promotes glycolysis and lactate production of colorectal cancer through regulating of miR-216b-5p/TRIM44 axis. Digestion. 2020;101(4):391–400. doi:10.1159/000500195
29. Li S, Guo W, Geng H, Wang C, Yang S, Xu X. LINC00511 exacerbated T-cell acute lymphoblastic leukemia via miR-195-5p/LRRK1 axis. Biosci Rep. 2020;40(5). doi:10.1042/BSR20193631
30. Huang M, Zheng J, Ren Y, et al. LINC00221 suppresses the malignancy of children acute lymphoblastic leukemia. Biosci Rep. 2020;40(5). doi:10.1042/BSR20194070
31. Hughes K, Watson CJ. The multifaceted role of STAT3 in mammary gland involution and breast cancer. Int J Mol Sci. 2018;19(6):1695. doi:10.3390/ijms19061695
32. Geiger JL, Grandis JR, Bauman JE. The STAT3 pathway as a therapeutic target in head and neck cancer: barriers and innovations. Oral Oncol. 2016;56:84–92. doi:10.1016/j.oraloncology.2015.11.022
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.