Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 16
Leu72Met Polymorphism in Ghrelin Gene: A Potential Risk Factor for Hypertension in Type 2 Diabetes Patients
Authors Buraczynska M, Golacki J, Zaluska W
Received 21 October 2022
Accepted for publication 9 February 2023
Published 1 March 2023 Volume 2023:16 Pages 557—564
DOI https://doi.org/10.2147/DMSO.S393373
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gian Paolo Fadini
Monika Buraczynska, Jakub Golacki, Wojciech Zaluska
Department of Nephrology, Medical University of Lublin, Lublin, Poland
Correspondence: Monika Buraczynska, Department of Nephrology, Medical University of Lublin, Jaczewskiego 8, Lublin, 20-950, Poland, Tel +48 81 7244716, Fax +48 81 7244357, Email [email protected]
Objective: Ghrelin (GHRL) is known to be engaged in metabolic and cardiovascular processes. There is evidence suggesting its involvement in the regulation of blood pressure and hypertension. The purpose of this preliminary case–control study was to determine the involvement of the Leu72Met (rs696217) polymorphism in the GHRL gene in type 2 diabetes (T2DM).
Methods: The Leu72Met polymorphism was genotyped in 820 individuals with T2DM and 400 healthy subjects by the PCR-RFLP technique. The polymorphism distribution was first compared in those withT2DM and controls, then in subgroups of participants representing different clinical phenotypes.
Results: No significant association was identified between Leu72Met and T2DM. The distribution of polymorphism was analyzed in subgroups of individuals with different clinical phenotypes (hypertension, diabetic nephropathy, obesity). In this analysis, rs696217 was associated with hypertension. The presence of T allele was associated with higher risk of hypertension (OR = 2.50, 95% CI 1.68– 3.73, p < 0.001). When adjusted for age, gender and BMI, the association was still significant (OR = 2.62, 95% CI 1.83– 3.96, p < 0.001). A post hoc power calculations based on a minor allele frequency revealed the power of 97% for comparison between HY+ and HY- subgroups.
Conclusion: This is the first study demonstrating that the ghrelin Leu72Met SNP is associated with hypertension in Caucasians with T2DM. If confirmed in larger studies in different populations, it may be a novel potential risk factor for hypertension in individuals withT2DM.
Keywords: ghrelin gene, type 2 diabetes, hypertension, Leu72Met polymorphism, risk allele
Introduction
Type 2 diabetes mellitus (T2DM) is a highly prevalent heterogeneous disorder of glucose metabolism, characterized by persistent hyperglycaemia. It accounts for almost 90% of all diabetes cases, causing the burden on affected people and the health care system.1 The disease is a multifactorial condition characterized by increased morbidity and mortality due to the development of macro- and microvascular complications.2 Multiple genetic variants and environmental factors, synergistically interacting with each other, play a role in the pathogenesis of type 2 diabetes and its complications.3 Investigating genetic risk factors for T2DM may improve our understanding of this disease and lead to a better diagnosis and treatment. One of the promising genes found to be associated with T2DM development is the gene encoding ghrelin peptide (GHRL) discovered by Kojima.4 This gastrointestinal peptide hormone is primarily synthesized in the stomach but also expressed in other tissues such as heart, kidney, and adipose tissue.5 Ghrelin is involved in accumulation of fat, gut motility and energy balance.6 It is also engaged in the blood glucose and insulin regulation. Ghrelin levels in plasma are associated with T2DM7–9 and metabolic syndrome (MetS).10 Two forms of ghrelin, acylated (AG) and deacylated (DAG), have different effects in patients with T2DM. AG levels are associated with elevated blood glucose levels and DAG levels show a strong negative association with excess body fat mass and insulin resistance.11
The GHRL gene is positioned on a short arm of chromosome 3 (3p25-26). It contains four exons encoding preproghrelin with 117 amino acids. Mature ghrelin peptide has 28 amino acids.12 Multiple polymorphisms in the ghrelin gene are located in the promoter and coding regions of the gene and some are associated with metabolic diseases.9 The polymorphism strongly associated with T2DM is the G to T change in the second exon of the gene (rs696217), replacing leucine at 72 amino acid with methionine (Leu72Met).13 Previously, this polymorphism was reported as associated with insulin metabolism, obesity, and metabolic syndrome.14–16 These conditions are closely linked to T2DM; thus, this led to investigations of GHRL Leu72Met variant in type 2 diabetes. The results of so far published studies from different populations are conflicting, with most of them reporting an association of Leu72Met with reduced risk of T2DM.13
This retrospective case–control study is aimed to assess the distribution of the Leu72Met variant in T2DM patients and healthy controls in order to analyze its role in type 2 diabetes.
Materials and Methods
Study Subjects
All individuals for this retrospective study were recruited from University Hospital and Polyclinic, Medical University of Lublin, between 2010 and 2019. Our study included a total of 820 unrelated adult participants with type 2 diabetes duration >10 years (mean age 65.2 ± 14.4 years). There were 446 (54%) men and 374 (46%) women in this cohort. All participants were Caucasians. Before enrolment in the study, a written informed consent was obtained from all participants, in accordance with standards of the Declaration of Helsinki (version 2013). Approval of the exact protocol of the study was obtained from the Ethics Committee of Medical University of Lublin (KE-0254/49).
Type 2 diabetes was diagnosed conforming to the guidelines of American Diabetes Association.1 The standard inclusion criteria were confirmed diagnosis of type 2 diabetes and age ≥30 years. A complete physical examination was performed on all patients, including fasting plasma glucose, glycated hemoglobin (HbA1c), full lipid profile, albumin-to-creatinine ratio (ACR), albumin excretion rate (AER) and body mass index (BMI). In addition to the classic symptoms of hyperglycemia (polyuria, polydipsia, loss of weight), the fasting plasma glucose level was >7 mmol/l or random level >11 mmol/l, and HbA1c level ≥6.5%. In this cross-sectional study, participants were not matched in terms of demographic or medical characteristics. Those excluded from the study were patients with T1DM, with other significant chronic diseases, eg, other endocrine disorders, haematological disorders, immunodeficiencies, pulmonary or rheumatological diseases, or malignancies were excluded from the study.
Cardiovascular disease was diagnosed in 603 individuals (73.5%). One or the combination of pathological phenotypes were diagnosed as cardiovascular disease: congestive heart failure, left ventricular hypertrophy, angina pectoris, ischemic heart disease, myocardial infarction, ischemic cerebral stroke. Its clinical manifestations were affirmed by relevant biochemical, radiographic, echocardiographic and vascular diagnostic criteria. In total, 582 participants (71%) were diagnosed with hypertension by the World Health Organization criteria, with average systolic blood pressure and diastolic blood pressure >140 mmHg and >90 mm Hg, respectively (the readings done on 2 different days).
Individuals in the control group (n = 400, mean age 57.5 ± 8.1 years), described earlier,17 were unrelated normoglycemic volunteers (blood donors and hospital staff members) who earlier underwent health examination. At the time of enrolment, they had no history of diabetes, cardiovascular disease or renal disorders. Those reporting the family history of these conditions in first-degree relatives were excluded. Individuals who did not sign an informed consent were also excluded. The control group was used for comparing Leu72Met genotype frequencies to values in subjects withT2DM.
Genotyping
Genomic DNA extraction from peripheral blood leukocytes was carried out by the standard procedure.18 Genotyping was carried out in order to determine the Leu72Met (rs696217) polymorphism genotypes in all patients and controls. The specific primer pair: forward 5’-TCTCTGGGGCTTCAGTCTTCT-3’ and reverse 5’-CACTGCCACCTCTCCTGC-3’ was used in the polymerase chain reaction-restricted fragment length polymorphism (PCR-RFLP) procedure to amplify 373 bp DNA fragment. Genomic DNA samples (200 ng) were amplified in 30 μL reactions. The initial denaturation step (95°C for 5 min) was followed by 30 cycles of denaturation (95°C), annealing (60°C) and extension (72°C) for 1 min each. The final extension (72°C) lasted 7 min. Ten µL of the PCR product were digested for 12 h at 37°C with 5 U of Bse NI (Bsr I) restriction enzyme (Thermo Fisher Scientific). Undigested reaction product and resulting digestion products were analyzed by electrophoresis in agarose gel (2.5%), stained with ethidium bromide. The length of products was 373 bp (no restriction site) for G allele and 271 + 101 bp for T allele. The blind DNA duplicates (96 samples) were used to control the correctness of genotyping. The rate of concordance was 100%. Additionally, automated sequencing was performed for 20 random samples for each genotype in the CEQ 8000 Genetic Analysis System (Beckman Coulter).
Statistical Analysis
Statistical calculations for study results were performed using Microsoft Excel and SPSS 18.0 package for Windows (SPSS, Inc., Chicago, IL, USA). For comparison of baseline characteristics between T2DM patients and controls, the continuous variables are reported as means ± SD. Categorical and qualitative variables are presented as numbers and percentages. The Hardy–Weinberg equilibrium (HWE) was tested separately in patients and controls, using the chi-square test. Distribution of genotypes and alleles was compared between groups and subgroups by means of chi-square test with 2×2 contingency and z statistics. For comparison of categorical and continuous variables between groups, we used Pearson’s Χ2 test of independence and t test for normally distributed variables and Mann–Whitney test for asymmetrically distributed. The odds ratios (OR) with 95% confidence intervals (CI) were calculated for the strength of observed associations. For association of genotypes with type 2 diabetes, we considered four genetic models: an additive, a co-dominant (TT or GT vs GG), a dominant (heterozygote with the minor allele homozygote: TT plus GT vs GG) and recessive model (the minor allele homozygote: TT vs GT plus GG). An interaction of alleles and genotypes of SNP with T2DM and clinical phenotype was evaluated with multivariate logistic regression analysis (ORs adjusted based on age, gender and BMI). For all tests, two-tailed type I error rate of 0.05 was considered statistically significant.
Results
Participants Characteristics
The genotypes of the rs696217 SNP in the GHRL gene were determined in 820 participants with T2DM and 400 healthy volunteers. The genotyping success rate reached 100%. The comparison of demographic, clinical and laboratory features of participants from both groups is presented in Table 1. The gender distribution was comparable between those with T2DM and controls (55% and 51.3% of the men, respectively, p = 0.211). People withT2DM were older and, as expected, showed significantly higher values in presented variables (BMI, systolic and diastolic blood pressure, total cholesterol and triglycerides) than control individuals (p < 0.001).
 |
Table 1 Demographic and Clinical Profile of Participants with T2DM and Healthy Controls |
Leu72Met Distribution in T2DM Patients Compared to Control Subjects
The results of genotyping are shown in Table 2. The observed genotype frequencies in our study were in agreement with values reported in other European studies.16,19 The genotype frequencies of rs696217 SNP were as predicted by Hardy–Weinberg equilibrium, in both T2DM patients and control group (p = 0.288 and p = 0.106, respectively). No statistically significant differences in genotype and allele distribution were detected between those withT2DM and healthy controls. There were also no significant differences observed between men and women (data not shown). Table 3 presents the distribution of the polymorphism in both groups according to the model of inheritance. When adjusted for age, gender and BMI, no significant association was observed in any of the models. The multiple logistic regression analysis was applied for assessment of the Leu72Met association with hypertension and possible interaction with other variables (Table 4). The Leu72Met polymorphism remained a significant risk predictor of hypertension in T2DM patients.
 |
Table 2 Genotype and Allele Distribution of Leu72Met Polymorphism in the GHRL Gene in Participants withT2DM and Healthy Individuals |
 |
Table 3 Distribution of the GHRL Gene Leu72Met Polymorphism According to the Model of Inheritance |
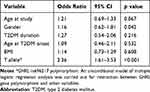 |
Table 4 The Results of Multivariate Logistic Regression Analysis |
Leu72Met Distribution in T2DM Patients with Different Clinical Phenotypes
We also analyzed the rs696217 polymorphism distribution in subgroups of participants with T2DM, with different clinical phenotypes (hypertension, diabetic nephropathy, obesity) (Table 5). In this analysis rs696217 was associated only with hypertension. The carriership of the T allele was associated with higher risk of hypertension (OR = 2.50,95% CI 1.68–3.73, p < 0.001). When adjusted for age, gender and BMI, the association was still significant (OR = 2.62, 95% CI 1.83–3.96, p < 0.001).
 |
Table 5 Genotype and Allele Distribution of Leu72Met Polymorphism in the GHRL Gene in Participants with T2DM with Different Clinical Phenotypes |
Discussion
Recently, the studies on genetic variants involved in human diseases become one of the most important areas in the pathogenesis of T2DM.20 The association between ghrelin gene polymorphisms and T2DM has been previously investigated in several studies covering different populations. A common Leu72Met polymorphism was reported to be associated with obesity, insulin metabolism and metabolic syndrome, the conditions linked to T2DM, hence the attempts to investigate its relationship with T2DM.21
Our preliminary study was designed to explore the involvement of the GHRL Leu72Met polymorphism in participants with type 2 diabetes mellitus. Two groups of individuals were studied, 820 individuals with T2DM and 400 healthy individuals as controls. Although the study population in this analysis was fairly large, we did not observe any association of Leu72Met SNP with type 2 diabetes. Previously, many studies on the association between Leu72Met polymorphism in the GHRL gene and T2DM have been reported, often with contradictory conclusions. Some studies observed the protective effect15,22–24 while in others Leu72Met increased the risk of T2DM.25 In the contrary, several studies documented, what is in agreement with our study, that Leu72Met variant is not associated with type 2 diabetes.26–30 In a Chinese study of 877 participants with T2DM and 864 controls, this polymorphism was not associated with diabetes but was involved in the etiology of insulin resistance.31 These controversies can, at least in part, be explained by differences in ethnicity, environmental factors or the design of experiments (selection and numbers of study subjects).
Through its expression in the heart and blood vessels, ghrelin is known to have a cardiovascular function and to be involved in the development of atherosclerosis. Intravenous injection of ghrelin in healthy individuals shows advantageous hemodynamic effects by lowering mean arterial pressure and increasing cardiac output. Low plasma ghrelin concentration would therefore have a detrimental hemodynamic effect.6,32 In a Finnish group of hypertensive people, low levels of plasma ghrelin were associated with hypertension.33
In the present study, we analysed the Leu72Met polymorphism distribution in subgroups of participants with T2DM with different clinical phenotypes. The results of this analysis unexpectedly indicated that the Leu72Met variant was associated with hypertension in those with T2DM. Carrying the T allele (72Met) was associated with higher risk of hypertension (OR = 2.50,95% CI 1.68–3.73, p < 0.001). Previously, another polymorphism in the GHRL gene, Arg51Gln, was found to be associated with hypertension. In the study of 1143 people with hypertension, the authors observed that carriers of the minor allele (Gln51) had a twofold greater risk of hypertension than noncarriers.34 In a Finnish study involving a cohort of 600 patients with hypertension, the Gln51 allele was more frequent in hypertensive subjects than in control ones (OR 2.63, 95% CI 1.37–5.08, p = 0.003).7 On the contrary, the Leu72Met SNP was not associated with hypertension or blood pressure.34 According to Ukkola et al, the obese female 72Met variant carriers had a lower prevalence of hypertension than the patients without it.35 In another Finnish study, Leu72Met polymorphism was associated with blood pressure levels, but not with hypertension, in a population of overweight individuals with impaired glucose tolerance.36 The discrepancy between our finding and those from other studies might reflect the genetic heterogeneity and be due in part to the sample size and/or phenotype of subjects. Our study involved 820 participants with type 2 diabetes lasting >10 years (without patients with impaired glucose tolerance in this cohort). In the Berthold et al study, among 2632 Caucasian participants, only 473 (18%) had type 2 diabetes.34 In a Finnish study out of 3004 subjects from 3 cohorts, 25% had type 2 diabetes (self-reported).35 The Mager et al study involved 522 overweight individuals with IGT.36 It is also possible that the effect of Leu72Met variant on the risk of hypertension in individuals with T2DM can depend on a set of particular environmental factors and specific conditions. It is also feasible that linkage disequilibrium with some other genes in the 3p26-p25 region might explain the observed Leu72Met association with hypertension.
It was demonstrated in earlier studies that in adults, the Met72Met genotype was associated with high ghrelin levels.33,35 The low level of ghrelin was independently associated with markedly increased BP level, insulin resistance and prevalence of T2DM. Also, the ghrelin levels in individuals with type 2 diabetes were lower than in those without diabetes.33 Joatar et al reported that they did not observe any statistically significant differences in plasma ghrelin levels between the carriers and noncarriers of the minor allele of GHRL Leu72Met polymorphism.30 Unfortunately, we did not quantify ghrelin levels in our study what significantly hinders the interpretation of observed association. Thus, the results from this study should be interpreted guardedly. The functional significance of Leu72Met polymorphism is not completely understood. According to some authors, this variant might affect the stability of mRNA or impede splicing of the preprohormone, thus affecting ghrelin secretion or activity.37 The exact biological mechanism underlying the association of ghrelin gene Leu72Met polymorphism and hypertension remains yet to be elucidated.
Conclusion
Our finding from this preliminary study suggests for the first time that the ghrelin Leu72Met polymorphism might be associated with increased risk of hypertension in Caucasians with type 2 diabetes mellitus. If these results are confirmed in larger studies and/or in different populations, it may be a novel potential risk factor for hypertension in people with T2DM.
Data Sharing Statement
The datasets used and analyzed during this study are available from the corresponding author on reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas, took part in drafting, revising or critically reviewing the article, gave final approval of the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Funding
This research did not receive any specific funding.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. American Diabetes Association. Standards of medical care in diabetes - 2019 abridged for primary care providers. Clin Diabetes. 2019;37:11–34. doi:10.2337/cd18-0105
2. Zheng Y, Ley SH, Hu FB. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14:88–98. doi:10.1038/nrendo.2017.151
3. O’Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: the end or the beginning? Science. 2005;307:370–373. doi:10.1126/science.1104346
4. Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi:10.1038/45230
5. Chen CY, Asakawa A, Fujimiya M, Lee SD, Inui A. Ghrelin gene products and the regulation of food intake and gut motility. Pharmacol Rev. 2009;61:430–481. doi:10.1124/pr.109.001958
6. Kojima M, Kanagawa K. Ghrelin: structure and function. Physiol Rev. 2005;85:495–522. doi:10.1152/physrev.00012.2004
7. Pöykkö S, Ukkola O, Kauma H, Savolainem MJ, Kesaniemi YA. Ghrelin Arg51Gln mutation is a risk factor for type 2 diabetes and hypertension in a random sample of middle-aged subjects. Diabetologia. 2003;46:455–458. doi:10.1007/s00125-003-1058-z
8. Al Qarni AA, Joatar FE, Das N, et al. Association of plasma ghrelin levels with insulin resistance in type 2 diabetes mellitus among Saudi subjects. Endocrinol Metab. 2017;32:230–240. doi:10.3803/EnM.2017.32.2.230
9. Poher AL, Tschöp MH, Müller TD. Ghrelin regulation of glucose metabolism. Peptides. 2018;100:236–242. doi:10.1016/j.peptides.2017.12.015
10. Ghalandari H, Hosseini-Esfahani F, Mirmiran P. The association of polymorphisms in leptin / leptin receptor gene and ghrelin / ghrelin receptor gene with overweight / obesity and the related metabolic disturbances: a review. Int J Endocrinol Metab. 2015;13:e19073. doi:10.5812/ijem.19073v2
11. Zang P, Yang CH, Liu J, et al. Relationship between acyl and desacyl ghrelin levels with insulin resistance and body fat mass in type 2 diabetes mellitus. Diabetes Metab Syndr Obes. 2022;15:2763–2770. doi:10.2147/DMSO.S368770
12. Kanamoto N, Akamizu T, Tagami T, et al. Genomic structure and characterization of the 5’ flanking region of the human ghrelin gene. Endocrinology. 2004;145:4144–4153. doi:10.1210/en.2003-1718
13. Liao N, Xie Z-K, Huang J, Xie Z-F. Association between the ghrelin Leu72Met polymorphism and type 2 diabetes risk: a meta-analysis. Gene. 2013;517:179–183. doi:10.1016/j.gene.2012.12.094
14. Xu LL, Xiang HD, Qiu CC, Xsu Q. Association of ghrelin polymorphisms with metabolic syndrome in Han nationality Chinese. Biomed Environ Sci. 2008;21:188–192. doi:10.1016/S0895-3988(08)60027-6
15. Zavarella S, Petrone A, Zampetti S, et al. A new variation in a promoter region, the −604 C>T, and the Leu72Met polymorphism of the ghrelin gene are associated with protection to insulin resistance. Int J Obes. 2008;32:663–668. doi:10.1038/sj.ijo.0803766
16. Vitolo E, Santini E, Seghieri M, et al. Heterozygosity for the rs696217 in the preproghrelin gene predicts weight loss after bariatric surgery in severe obese individuals. Obes Surg. 2017;27:961–967. doi:10.1007/s11695-016-2387-6
17. Buraczynska M, Gwiazda-Tyndel K, Drop B, Zaluska W. Renalase gene Glu37Asp polymorphism affects susceptibility to diabetic retinopathy in type 2 diabetes mellitus. Acta Diabetol. 2021;58:1596–1602. doi:10.1007/s00592-021-01740-8
18. Madisen L, Hoar DI, Helroyd CD, Crisp H, Hodes ME. DNA banking: the effect of storage of blood and isolated DNA on the integrity of DNA. American Journal of Medical Genetics. 1987;27:379–390. doi:10.1002/ajmg.1320270216
19. Hansson C, Annerbrink K, Nilsson S, et al. A possible association between panic disorder and the polymorphism in the preproghrelin gene. Psychiatry Res. 2013;206:22–25. doi:10.1016/j.psychres.2012.09.051
20. Fuchsberger C, Funnick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi:10.1038/nature18642
21. Li Y, Lu X, Yang X, et al. GHRL gene Leu72Met polymorphism and type 2 diabetes mellitus: a meta-analysis involving 8194 participants. Front Endocrinol. 2019;10:article559. doi:10.3369/endo.2019.00559
22. Mager U, Lindt V, Lindström J, et al. Association of the Leu72Met polymorphism of the ghrelin gene with the risk of type 2 diabetes in subjects with impaired glucose tolerance in the Finnish Diabetes Prevention Study. Diabetic Med. 2006;23:685–689. doi:10.1111/j.1464-5491.2006.01870.x
23. Zhuang L, Li M. Yu et al. The Leu72Met polymorphism of the GHRL gene prevents the development of diabetic nephropathy in Chinese patients with type 2 diabetes mellitus. Mol Cell Biochem. 2014;387:19–25. doi:10.1007/s11010-013-1865-6
24. Rivera-Leon EA, Llamas-Covarrubias MA, Sanchez-Enriquez S, et al. Leu72Met polymorphism of GHRL gene decreases susceptibility to type 2 diabetes mellitus in a Mexican population. BMC Endocrinol Dis. 2020;20:109. doi:10.1186/s12902-020-00596-3
25. Takezawa J, Yamada K, Morita A, Alba N, Watanabe S. Preproghrelin gene polymorphisms in obese Japanese: association with diabetes mellitus in men and with metabolic syndrome parameters in women. Obesity Res Clin Pract. 2009;3:179–191. doi:10.1016/j.orcp.2009.04.003
26. Larsen LH, Gjesing AP, Sorensen TA, et al. Mutation analysis of the preproghrelin gene: no association with obesity and type 2 diabetes. Clin Biochem. 2005;38:420–424. doi:10.1016/j.clinbiochem.2005.01.008
27. Kim SY, Jo DS, Hwang PH, Park SK, Yi HK, Lee DY. Preproghrelin Leu72Met polymorphism is not associated with type 2 diabetes mellitus. Metabolism. 2006;55:366–370. doi:10.1016/j.metabol.2005.09.011
28. Garcia EA, King P, Sidhu K, et al. The role of ghrelin and ghrelin-receptor gene variants and promoter activity in type 2 diabetes. Eur J Endocrinol. 2009;161:307–315. doi:10.1530/EJE-09-0122
29. Hedayatizadeh-Omran A, Rafei A, Khajavi R, Alizadeh Navaei R, Mokhberi V, Moradzadeh K. Association between ghrelin gene (Leu72Met) polymorphism and ghrelin serum level with coronary artery disease. DNA Cell Biol. 2014;33. doi:10.1089/dna2013.2218
30. Joatar FE, AlQarni AA, Ali ME, et al. Leu72Met and other intronic polymorphisms in the GHRL and GHDR genes are not associated with type 2 diabetes mellitus, insulin resistance, or serum ghrelin levels in a Saudi population. Endocrinol Metab. 2017;32:360–369. doi:10.3803/EnM.2017.32.3.360
31. Liu J, Liu J, Tian L, et al. Association of ghrelin Leu72Met polymorphism with type 2 diabetes mellitus in Chinese population. Gene. 2012;504:309–312. doi:10.1016/j.gene.2012.03.025
32. Nagaya N, Kojima M, Uematsu M, et al. Hemodynamic and hormonal effects of human ghrelin in healthy volunteers. Am J Physiol Regul Integr Comp Physiol. 2001;280:R1483–R1487. doi:10.1152/ajpregu.2001.280.5.R1483
33. Pöykkö SM, Kellokoski E, Hörkkö S, Kauma H, Kesäniemi YA, Ukkola O. Low plasma ghrelin is associated with insulin resistance, hypertension and the prevalence of type 2 diabetes. Diabetes. 2003;52:2546–2553. doi:10.2337/diabetes.52.10.2546
34. Berthold HK, Giannakidou E, Krone W, Gouni-Berthold I. Influence of ghrelin gene polymorphism on hypertension and atherosclerotic disease. Hypertens Res. 2010;33:155–160. doi:10.1038/hr.2009.194
35. Ukkola O, Ravussin E, Jacobson P, et al. Role of ghrelin polymorphisms in obesity based on three different studies. Obes Res. 2002;10:782–791. doi:10.1038/oby.2002.106
36. Mager U, Kolehmainen M, Lindström J, et al. Association between ghrelin gene variations and blood pressure in subjects with impaired glucose tolerance. Am J Hypertens. 2006;19:920–926. doi:10.1016/j.amjhyper.2006.02.017
37. Ukkola O. Genetic variants of ghrelin in metabolic disorders. Peptides. 2011;32:2319–2322. doi:10.1016/j.peptides.2011.04.013
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
