Back to Journals » Cancer Management and Research » Volume 10
Knockdown of PKM2 suppresses tumor progression in human cervical cancer by modulating epithelial–mesenchymal transition via Wnt/β-catenin signaling
Authors Lin Y, Meng F, Lu Z, Chen K, Tao Y, Ouyang Y, Cao X
Received 25 June 2018
Accepted for publication 9 August 2018
Published 4 October 2018 Volume 2018:10 Pages 4191—4202
DOI https://doi.org/10.2147/CMAR.S178219
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yanzhu Lin,1,* Fanqing Meng,2,* Zhiyuan Lu,3 Kai Chen,1 Yalan Tao,1 Yi Ouyang,1 Xinping Cao1
1Department of Radiation Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China; 2Department of Anesthesiology, Jinan Maternity and Child Care Hospital, Jinan, China; 3Department of Oral and Maxillofacial Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
*These authors contributed equally to this work
Background: Pyruvate kinase isozyme type M2 (PKM2) is a key glycolytic enzyme and is upregulated in multiple human malignancies. However, the role of PKM2 in human cervical cancer (CC) remains elusive. Thus, this study explored the role of PKM2 in CC by detecting its expression patterns in human CC tissues and cell lines and investigated its effects on cell proliferation and invasion.
Materials and methods: Quantitative reverse transcription polymerase chain reaction (qRT-PCR), immunohistochemistry and western blotting assays were used to detect the expression of PKM2 in CC tissues and CC cells. In vitro, we overexpressed and knocked down PKM2 expression in CC cell lines and investigated the biological function and underlying mechanism of PKM2 in cervical carcinogenesis.
Results: The results showed that PKM2 mRNA and protein were highly expressed in CC tissues and cell lines. Furthermore, increasing PKM2 expression was closely correlated with the clinical stage (P=0.001) and lymph node metastasis (P=0.023). The functional roles of PKM2 were determined using Cell Counting Kit-8, colony formation, and transwell assays. The results showed that PKM2 knockdown inhibited cell proliferation and the migratory and invasive capacities of CC cells, suppressed epithelial–mesenchymal transition (EMT), and inhibited Wnt/β-catenin signaling in vitro. However, overexpression of PKM2 led to increased proliferation and invasion activity as well as the EMT in CC cells.
Conclusion: Taken together, our study results revealed that PKM2 may act as a molecular target for CC treatment.
Keywords: Cervical cancer, PKM2, invasion, epithelial–mesenchymal transition, migration
Introduction
Cervical cancer (CC) is the second most common fatal gynecological malignancy worldwide.1 Despite advancements in radiotherapy and surgical therapy, numerous patients with late-stage CC still experience metastasis and recurrence, particularly in developing countries.2 Hence, efforts are being made to develop effective targeted therapies for CC.
Tumor cells undergo glycolysis even under highly oxygenated conditions, consuming a large amounts of glucose and generating lactate, a phenomenon termed aerobic glycolysis or the Warburg effect.3 Pyruvate kinase isozyme type M2 (PKM2) is an isozyme of pyruvate kinase that functions as a rate-limiting enzyme catalyzing the conversion of phosphoenolpyruvate and adenosine diphosphate to pyruvate in the final step of the glycolytic pathway.4 PKM2 is overexpressed in several malignancies, including CC and is associated with tumor development and poor prognosis.5,6 Landt et al7 reported that PKM2 can be used as a biomarker for differentiating between malignant and premalignant cervical lesions. Furthermore, a study showed that positive PKM2 expression indicated the development of cervical carcinoma.8 However, the function of PKM2 in the tumorigenesis and progression of CC is yet to be explored in detail.
The epithelial–mesenchymal transition (EMT) is a biological process through which epithelial cells lose their polarity and become mesenchymal cells, gaining migratory properties that increase invasiveness in cancer progression.9,10 Growing evidence suggests that the EMT is associated with metastasis and occurs during the development of various carcinomas, including CC.11 Numerous signaling pathways are involved in the EMT process, including the Wnt, Notch, nuclear factor-kappa B, TGF-β, and RTK/Ras.12 Among them, Wnt/β-catenin is essential for the regulation of cell growth and development; its abnormal activation is common in human cancers, including CC.13 In the Wnt pathway, inactivation of a destruction complex targeting β-catenin for proteasomal degradation results in the translocation of β-catenin to the nucleus, which activates the downstream target oncogenes of β-catenin, such as c-Myc and cyclin D1.
To determine the expression pattern of PKM2 in CC, we profiled the expression status of PKM2 in CC tissues. Moreover, we analyzed the association between the expression of PKM2 and the clinicopathological characteristics of CC. After examining the mechanisms underlying the association between PKM2 and metastasis, we found that Wnt/β-catenin signaling was involved in PKM2-mediated invasion.
Materials and methods
Tissue samples
Between June 2016 and December 2017, a total of 103 cervical squamous cell carcinoma (SCC) specimens were collected through biopsy from patients who were not subjected to preoperative radiotherapy or chemotherapy at Sun Yat-sen University Cancer Center. Fifteen normal cervical epithelial tissue samples were obtained from patients without CC through surgical resection at the same center. A total of 36 paired CC tissues and adjacent nontumor tissues were randomly selected for quantitative polymerase chain reaction (qPCR), and seven paired tissues were selected for Western blot analysis. All CC and normal tissues were confirmed by pathological diagnosis. The clinicopathological information collected included age, tumor size, differentiation grade, lymph node metastasis, and International Federation of Gynecology and Obstetrics (FIGO) stage. All patients provided written informed consent, and the study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center.
Immunohistochemistry
Immunohistochemical analysis was performed to measure PKM2 protein expression. In brief, samples were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned, and processed for immunohistochemical staining. Slides were baked at 60°C for 1 hour, followed by deparaffinization with xylene and rehydration. Subsequently, they were treated with 3% hydrogen peroxide to quench the endogenous peroxidase activity, followed by incubation with 5% bovine serum albumin to block nonspecific binding. Sections were incubated with anti-PKM2 (1:150 dilution, cell-signalling technology [CST]) overnight at 4°C. After washing, the tissue sections were treated with a secondary antibody, followed by incubation with conjugated rabbit peroxidase streptavidin. Color was developed using a DAB (3,3’-diaminobenzidine) Horseradish Peroxidase Color Development Kit (Fuzhou Maixin Biotech Inc., Fuzhou, China). Subsequently, the tissue sections were then counterstained with hematoxylin, dehydrated, and mounted. Finally, the sections were viewed under a bright-field microscope. The tissue sections subjected to immunohistochemical staining for PKM2 were reviewed and scored separately by two pathologists blinded to clinical parameters. Any disagreements were arbitrated by a third pathologist. To assess PKM2 expression, staining intensity was scored as negative =0; weak =1; moderate =2; and strong =3, and staining extent was scored as 1%–25%=1; 26%–50%%=2; 51%–75%%=3; and >76%=4. The final score was calculated by multiplying these two variables. If the total score was >7, it was defined as high expression, or if not, it was defined as low expression.
Cell culture
The normal cervical epithelial ECT cell line and the human CC cell lines HeLa, SiHa, Caski, MS751, HeLa229, and C33A were obtained from American Type Culture Collection (Manassas, VA, USA). SiHa, MS751, and HeLa229 cells were cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and penicillin–streptomycin, whereas other cell lines were cultured in Roswell Park Memorial Institute-1640 medium containing 10% FBS at 37°C in a humidified tissue culture incubator under 5% CO2.
Real-time (RT) qPCR
Total RNA from cells or tissues was extracted using TRI reagent (TRIzol, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol, and 500 ng of RNA was used to obtain cDNA through reverse-transcription by using PrimeScript RTMasterMix (Takara Bio, Kusatsu, Japan). A quantitative RT (qRT)-PCR reaction was conducted using a SYBR Green PCR Master Mix (Takara Bio) on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Primers used in this study are listed in Table S1. Relative expression was determined by normalizing to the expression of β-actin. Relative gene expression levels were detected and calculated using the ΔΔCt comparative method.
Western blot assay
Total proteins were extracted as previously described.14 Tissues and cells were lysed in RIPA buffer (Beyotime, China), and protein concentrations were measured using the bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). Equal protein extracts were run on SDS-PAGE and subsequently transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). After the membrane was blocked for 1 hour using 5% skim milk, it was incubated with a primary antibody overnight at 4°C. Finally, the membrane was incubated with their respective horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 2 hours. The sources of the antibodies and their concentrations are as follows: rabbit anti-PKM2 (1:1000, CST), rabbit anti-c-Myc (1:1000, CST), mouse anti-E-cadherin (1:1000, BD), mouse anti-vimentin (1:1000, BD), mouse anti-β-catenin (1:1000, Santa Cruz Biotechnology Inc.), rabbit anti-slug (1:1000, CST), rabbit anti-snail (1:1000, CST), rabbit anti-cycline D1 (1:1000, CST), and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (1:1000, CST). The protein bands of interest were visualized using enhanced chemiluminescence reagents (Millipore, Burlington, MA, USA).
Generation of stable PKM2 knockdown or overexpression cells
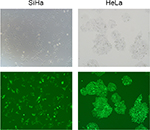
Enhanced green fluorescent protein-tagged shRNA against PKM2 (shPKM2) and PKM2-overexpression lentivirus were purchased from Shanghai Genechem Co., Ltd; the sequences are shown in Table S2. In brief, cells were grown to 50%–60% confluence and infected with lentivirus according to the manufacturer’s instructions. After 12 hours infection, the virus-containing medium was removed, and stable clones were selected with 8 µg/mL of puromycin for 48 hours.
Colony formation assay
For colony formation, the indicated cells were plated at 200 cells/well and incubated for 14 days to form colonies. The colonies were fixed using methanol and stained with 0.5% crystal violet for 15 minutes; and colonies of >50 cells were counted under a light microscope.
Transwell assay
Migration and invasion were assessed using Transwell plates. For the invasion assay, Matrigel chambers (BD Biosciences, San Jose, CA, USA) were used to determine the effect of cells on invasion according to the manufacturer’s instructions. The indicated cells at a density of 5×104 cells/well were resuspended in 200 µL into the upper chamber (8-µm pore size, Costar, Corning, NY, USA) of a Transwell system, whereas the lower chamber was filled with 0.5 mL of medium supplemented with 10% FBS. After incubation for 24 hours at 37°C, the invasive cells were fixed with 100% methanol and stained with 0.5% crystal violet before counting under an inverted microscope. For the migration assay, the indicated cells were plated on uncoated Matrigel upper chambers. The transmembraned cells were estimated under a microscope (Nikon, Tokyo, Japan) at 200× magnification.
Cell Counting Kit-8 (CCK-8) assay
The indicated cells were seeded into 96-well plates at density of 3×103 cells per well. Cell viability was determined using CCK-8 (Dojindo, Kumamoto, Japan) every 24 hours and by measuring absorbance at 450 nm using an ELx800 plate reader (BioTek, Winooski, VT, USA) following the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using SPSS version 18.0 (SPSS Inc, Chicago, IL, USA). Data are presented as the mean ± SD. The chi-squared test was used to analyze clinicopathological characteristics. Comparisons between two groups were evaluated using Student’s t-test. Differences among the groups were tested using a one-way ANOVA. All experiments were repeated at least three times. A P value of<0.05 was considered statistically significant.
Results
Expression of PKM2 is significantly upregulated in CC
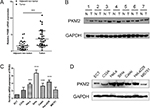
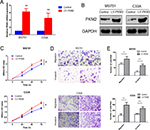
To evaluate the expression status of PKM2 in human CC tissues, the mRNA expression level of PKM2 of 36-paired CC and adjacent noncancerous tissues were examined through qRT-PCR. The mRNA expression level of PKM2 was upregulated in CC tissues than in adjacent noncancerous ones (Figure 1A; P<0.001). We also found that the PKM2 protein level expression was higher in CC tissues than in adjacent noncancerous ones (Figure 1B). Accordingly, PKM2 expression was further examined in six CC cell lines and one normal epithelial cell line through qRT-PCR and Western blot. The results revealed that PKM2 expression was considerably higher in CC cell lines than in normal epithelial cell line (Figure 1C, D). These results indicated that both mRNA and protein levels of PKM2 were significantly increased in CC.
Correlations between PKM2 expression and clinicopathological parameters in CC
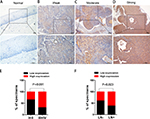
To determine the clinical significance of PKM2 in the development and progression of CC, an immunohistochemical assay was performed. Representative photographs of PKM2 expression are shown in Figure 2, where the expression of PKM2 can be observed to exhibit strong staining in cervical SCC and was predominantly localized in the cytoplasm, whereas the expression of PKM2 in normal cervical epithelial tissues were negative, indicating that PKM2 was upregulated in CC. The relationships between PKM2 expression and the clinicopathological factors of CC patients are shown in Table 1. Our results demonstrated that PKM2 expression was positively correlated with the FIGO stage (P=0.001) and lymph node metastasis (P=0.023). However, no significant difference existed between PKM2 expression and tumor size, differentiation, and age. Furthermore, we evaluated the prognostic effect of PKM2 expression by using Kaplan–Meier Plotter analysis in the publicly available web-based tool GEPIA (http://gepia.cancer-pku.cn/) based on the TCGA and GTEx databases. The Kaplan–Meier plots of overall survival (OS; Figure S1) indicated that patients with high expression of PKM2 mRNA were associated with lower OS (P=0.015), whereas no significant difference existed in disease-free survival in CC (P=0.74). These data indicate that PKM2 is associated with the progression and poor prognosis of CC.
  | Table 1 Correlation between PKM2 expression and clinicopathological parameters for cervical carcinoma Note: *P<0.05, **P<0.01. Abbreviation: PKM2, pyruvate kinase isozyme type M2. |
Effect of PKM2 on the cell proliferation of CC in vitro
To investigate the biological roles of PKM2 in CC progression, HeLa and SiHa cells were transfected with lentivirus shRNA vectors containing nontargeting shRNA (Control) or shRNA against PKM2 as models to generate stable cell lines. Western blotting and qPCR analysis showed that the PKM2 knockdown dramatically reduced PKM2 expression compared with the Control group (Figure 3A, B, Figure S2). Next, we examined the cell proliferative capacity of PKM2 on CC cells by using CCK-8 and colony formation assays. As shown in Figure 3C–F results indicated that the suppression of PKM2 significantly inhibited cell proliferation and reduced cell viability compared with the Control group in both cell lines (P<0.01 and P<0.05). To further confirm the role of PKM2 in cell proliferation, we selected the MS751 and C33A cell lines with low PKM2 expression to construct the PKM2 overexpression model (LV-PKM2) and evaluated their cell viability. PKM2 was successfully overexpressed in both the cell lines (Figure S3A, B), and the CCK8 assay showed that the overexpression of PKM2 significantly enhanced cell proliferation (Figure S3C). Collectively, our data implied that PKM2 affected proliferation in CC cells.
Knockdown of PKM2 suppresses CC migration and invasion in vitro
CC is characterized by a high metastasis potential in clinical practice. Subsequently, we determined whether PKM2 affects the cell migratory and invasive capacities of CC cells. The results of the transwell assay showed that knockdown of PKM2 markedly reduced the migration and invasion ability of HeLa and SiHa cells. Representative images and quantitative graphs representing the cell count are shown in Figure 4. However, this result was in opposition to that of C33A and MS751 cells between the overexpression and Control groups (Figure S3D, E). The results suggested that PKM2 plays a significant role in cell migration and invasion in CC cells.
PKM2 regulates the expression of EMT markers through Wnt/β-catenin signaling
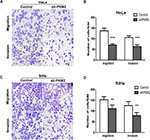
EMT is associated with CC.15 To investigate the effect of PKM2 on EMT in CC, we examined the expression of classic epithelial (E-cadherin) and mesenchymal (vimentin, Snail, and Slug) markers after PKM2 downregulation in CC cells. As shown in Figure 5A, B, decreased levels of the mesenchymal markers vimentin, Snail, and Slug were observed in the PKM2-knockdown CC cells, while the epithelial marker E-cadherin was significantly upregulated in SiHa cells, and barely detected in HeLa cells. However, the expression of E-cad was downregulated and those of the EMT markers vimentin, Snail, and Slug were upregulated in the PKM2 overexpression group of both cell lines. These results demonstrated that PKM2 knockdown potentially inhibited EMT in CC cells.
The Wnt/β-catenin pathway plays a crucial role in cell proliferation and promotes EMT in cancer development. To examine whether PKM2 regulates the activity of the Wnt/β-catenin pathway, we examined the related protein expression level. Compared with the Control group, knockdown of PKM2 significantly reduced β-catenin, c-Myc, and cyclin D1 expression levels in both cell lines. PKM2 overexpression in MS751 and C33A cells led to an opposite effect. Collectively, these data indicate that Wnt/β-catenin signaling has a crucial role in PKM2-induced CC cell migration and invasion.
Discussion
PKM2 is upregulated and acts as a tumor promotor in various human cancers.10,16 Consistently, in the current study, we investigated the role of PKM2 in CC and determined the underlying mechanism. We found that PKM2 expression was remarkably increased in CC tissues and cell lines. Furthermore, we verified that PKM2 facilitated the EMT process through the Wnt/β-catenin pathway to promote cell proliferation and invasion in CC. Therefore, our data suggest that PKM2 may be a novel biomarker for CC therapy.
Aerobic glycolysis is widely accepted as a hallmark of invasive cancers; thus, regulation of aerobic glycolysis has become a popular research topic in recent years.17 PKM2 is one of the four isozymes of PK (M1, M2, L, and R) that is mainly expressed in embryonic tissues and proliferative cells, especially in cancer cells.18 PKM2 is a step-limiting member of the glycolytic pathway, and emerging evidence indicates that PKM2 is closely associated with the onset and development of multiple tumors. For example, aberrant expression of PKM2 in esophageal SCC,19 hepatocellular carcinoma,20 and colorectal cancer21 has been associated with aggressive disease, poor differentiation, metastases development, and poor clinical prognosis. Nevertheless, few studies have explored the function of PKM2 in detail in the onset and development of CC. In this study, we determined the expression of PKM2 in human CC tissues and investigated its possible roles. Specifically, PKM2 expression was significantly increased in CC tissues, and the strong positive staining in CC tissue samples further supported the results. Additionally, PKM2 upregulation was positively correlated with the FIGO stage and lymph node metastasis, suggesting that PKM2 plays a crucial role in CC development. In addition, the publicly available web-based tool GEPIA showed that high PKM2 expression predicted poor outcomes, which implied that PKM2 may serve as an independent prognostic indicator of CC.
To further explore the biological effect of PKM2 on CC cells, we investigated the effect of PKM2 knockdown on the proliferation, invasion, and migration of two CC cell lines. First, we examined the expression level of PKM2 in different CC cell lines. Our results showed that PKM2 expression was upregulated in CC cell lines. We selected HeLa and SiHa cell lines transfected with shRNA of PKM2, which indicated high PKM2 expression as the cell model. The findings of in vitro experiments showed that the knockdown of PKM2 inhibited CC cell proliferation, migration, and invasion. By contrast, the overexpression of PKM2 promoted cell proliferation and invasion, which was consistent with the results of a previous study.22
Although the role of PKM2 in cell migration and invasion has been described extensively, its mechanisms remain unclear. EMT is a biological process through which epithelial cells lose their polarity, which promotes the malignancy in tumor progression. Loss of E-cadherin and acquisition of vimentin are two critical steps in EMT.23 The members of the Snail family, Snail and Slug, play a critical role in EMT during embryonic development and tumor progression. Emerging research has suggested that PKM2 plays a role in tumorigenesis through EMT. One study reported that PKM2 functions as an interacting partner of epidermal growth factor receptor (EGFR) and induces the EMT phenotype in colon cancer.24 Furthermore, STAT3 activation is closely related to PKM2. Yang et al25 reported that overexpression of PKM2 promoted colorectal cancer cell migration and cell adhesion by inducing EMT through the STAT3 signaling pathway. In the current study, we analyzed the expression patterns of relevant genes involved in EMT, including E-cadherin, vimentin, Snail, and Slug. The transcription factors Snail and Slug were significantly repressed after knockdown of PKM2 in HeLa and SiHa cells, whereas the epithelial marker E-cadherin was upregulated. By contrast, overexpression of PKM2 promoted EMT, indicating that PKM2 plays a crucial role in EMT in CC. These data suggest that PKM2 knockdown suppressed tumor metastasis by preventing the EMT process in CC cells.
Abnormal activation of the Wnt/β-catenin signaling pathway plays a crucial role in PKM2-related proliferation and invasion. Yang et al26 demonstrated that EGFR activation induces K433 of PKM2 to bind to c-Src-phosphorylated Y333 of β-catenin, leading to histone H3 acetylation and cyclin D1 expression and further promoting tumor cell proliferation and brain tumor development. Furthermore, Wu et al27 found that PKM2 depletion provokes glutamine metabolism by enhancing the β-catenin signaling pathway, thereby activating its downstream c-Myc-mediated glutamine metabolism in colon cancer. Another study found that CCs overexpressed β-catenin through DNA methylation of the promoter region.28 One study showed that PKM2 affected the β-catenin signaling pathway through miR-200a.29 These studies indicate that PKM2 is concerned with the regulation of the Wnt/β-catenin signaling pathway. Nuclear β-catenin is believed to be the hallmark of activation of the canonical Wnt pathway. We further examined the expression of two β-catenin downstream genes, c-Myc and cyclin D1, to clarify the effects of PKM2 expression on the Wnt pathway in CC. We found that PKM2 suppression significantly reduced the β-catenin level, and c-Myc and cyclin D1 levels in vitro, whereas PKM2 overexpression increased cyclinD-1 and c-Myc levels as well as β-catenin expression, indicating that PKM2 may participate in regulating the Wnt pathway in CC. These data suggest that knockdown of PKM2 inhibited the proliferation and invasion of human CC cells in part through suppressing Wnt/β-catenin signaling pathway activation. However, more research is required to clarify the role of Wnt/β-catenin in PKM2-related proliferation and invasion in CC.
Overall, our study confirmed the positive expression of PKM2 in human CC tissues and cell lines, as well as demonstrated that PKM2 promotes CC cell migration and invasion by inducing EMT through the Wnt/β-catenin signaling pathway. However, future studies should collect a large sample size of CC tissues and analyze the relationship between PKM2 expression and the prognosis of CC patients. A more comprehensive investigation will be conducted in vivo.
Acknowledgments
We gratefully thank two pathologists Lili Liu and Rongzhen Luo for their assistance in specimens evaluation. This work was supported by the Guangdong Natural Science Foundation (2017A030310197). The authenticity of this article was validated by uploading the key raw data into the Research Data Deposit public platform (www.researchdata.org.cn) with the approval number RDDB2018000381.
Disclosure
The authors report no conflicts of interest in this work.
References
Matoda M, Takeshima N, Michimae H, et al. Postoperative chemotherapy for node-positive cervical cancer: results of a multicenter phase II trial (JGOG1067. Gynecol Oncol. 2018;149(3):513–519. | ||
Denny L, de Sanjose S, Mutebi M, et al. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet. 2017;389(10071):861–870. | ||
Yang L, Zhang W, Wang Y, et al. Hypoxia-induced miR-214 expression promotes tumour cell proliferation and migration by enhancing the Warburg effect in gastric carcinoma cells. Cancer Lett. 2018;414:44–56. | ||
Zhou Z, Li M, Zhang L et al. Oncogenic kinase-induced PKM2 tyrosine 105 phosphorylation converts non-oncogenic PKM2 to a tumor promoter and induces cancer stem-like cells. Cancer Res. 2018;78(9):2248–2261. | ||
Dhar DK, Olde Damink SW, Brindley JH, et al. Pyruvate kinase M2 is a novel diagnostic marker and predicts tumor progression in human biliary tract cancer. Cancer. 2013;119(3):575–585. | ||
Hathurusinghe HR, Goonetilleke KS, Siriwardena AK. Current status of tumor M2 pyruvate kinase (tumor M2-PK) as a biomarker of gastrointestinal malignancy. Ann Surg Oncol. 2007;14(10):2714–2720. | ||
Landt S, Jeschke S, Koeninger A, et al. Tumor-specific correlation of tumor M2 pyruvate kinase in pre-invasive, invasive and recurrent cervical cancer. Anticancer Res. 2010;30(2):375–381. | ||
Yuan Y, Guo-Qing P, Yan T, Hong-Lin Y, Gong-Hua H, Cai-Gao Z. A study of PKM2, PFK-1, and ANT1 expressions in cervical biopsy tissues in China. Med Oncol. 2012;29(4):2904–2910. | ||
Zhai D, Cui C, Xie L, Cai L, Yu J. Sterol regulatory element-binding protein 1 cooperates with c-Myc to promote epithelial-mesenchymal transition in colorectal cancer. Oncol Lett. 2018;15(4):5959–5965. | ||
Lu Z, Guo H, Lin Y, Shen L, Yin C, Xie S. Effects of PTEN gene silencing on invasion and EMT in oral squamous carcinoma Tca8113 cells. J Oral Pathol Med. 2017;46(1):31–38. | ||
Ammothumkandy A, Maliekal TT, Bose MV, et al. CD66 and CD49f expressing cells are associated with distinct neoplastic phenotypes and progression in human cervical cancer. Eur J Cancer. 2016;60:166–178. | ||
Pastushenko I, Brisebarre A, Sifrim A, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–468. | ||
Bahrami A, Hasanzadeh M, Shahidsales S, et al. Clinical significance and prognosis value of Wnt signaling pathway in cervical cancer. J Cell Biochem. 2017;118(10):3028–3033. | ||
Lu Z, Lin Y, Peng B, Bao Z, Niu K, Gong J. Hydrogen-rich saline ameliorates hepatic ischemia-reperfusion injury through regulation of endoplasmic reticulum stress and apoptosis. Dig Dis Sci. 2017;62(12):3479–3486. | ||
Xiong Y, Sun F, Dong P, et al. iASPP induces EMT and cisplatin resistance in human cervical cancer through miR-20a-FBXL5/BTG3 signaling. J Exp Clin Cancer Res. 2017;36(1):48. | ||
Zhang X, He C, He C, et al. Nuclear PKM2 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Pathol Res Pract. 2013;209(8):510–515. | ||
Avanzato D, Pupo E, Ducano N, et al. High USP6NL levels in breast cancer sustain chronic AKT phosphorylation and GLUT1 stability fueling aerobic glycolysis. Cancer Res. 2018;78(13):3432–3444. | ||
Christofk HR, vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. | ||
Zhan C, Shi Y, Lu C, Wang Q. Pyruvate kinase M2 is highly correlated with the differentiation and the prognosis of esophageal squamous cell cancer. Dis Esophagus. 2013;26(7):746–753. | ||
Fan F, Wu H, Liu Z, et al. Nuclear PKM2 expression, an independent risk factor for ER after curative resection of hepatocellular carcinoma. Biomed Pharmacother. 2016;84:1858–1864. | ||
Ewald N, Schaller M, Bayer M, et al. Fecal pyruvate kinase-M2 (tumor M2-PK) measurement: a new screening concept for colorectal cancer. Anticancer Res. 2007;27(4A):1949–1952. | ||
Yan XL, Zhang XB, Ao R, Guan L. Effects of shRNA-mediated silencing of PKM2 gene on aerobic glycolysis, cell migration, cell invasion, and apoptosis in colorectal cancer cells. J Cell Biochem. 2017;118(12):4792–4803. | ||
Lu Z, Guo H, Lin Y, Shen L, Yin C, Xie S. Effects of PTEN gene silencing on invasion and EMT in oral squamous carcinoma Tca8113 cells. J Oral Pathol Med. 2017;46(1):31–38. | ||
Shi Y, Liu N, Lai W, et al. Nuclear EGFR-PKM2 axis induces cancer stem cell-like characteristics in irradiation-resistant cells. Cancer Lett. 2018;422:81–93. | ||
Yang P, Li Z, Fu R, Wu H, Li Z. Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell Signal. 2014;26(9):1853–1862. | ||
Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. | ||
Wu H, Li Z, Yang P, Zhang L, Fan Y, Li Z. PKM2 depletion induces the compensation of glutaminolysis through β-catenin/c-Myc pathway in tumor cells. Cell Signal. 2014;26(11):2397–2405. | ||
Shinohara A, Yokoyama Y, Wan X, et al. Cytoplasmic/nuclear expression without mutation of exon 3 of the beta-catenin gene is frequent in the development of the neoplasm of the uterine cervix. Gynecol Oncol. 2001;82(3):450–455. | ||
Wu H, Li Z, Yang P, Zhang L, Fan Y, Li Z. PKM2 depletion induces the compensation of glutaminolysis through β-catenin/c-Myc pathway in tumor cells. Cell Signal. 2014;26(11):2397–2405. |
Supplementary materials
  | Table S1 The primers used in qRT-PCR analysis Abbreviation: PKM2, pyruvate kinase isozyme type M2; qRT-PCR, quantitative real-time polymerase chain reaction; shPKM2, shRNA against PKM2. |
  | Table S2 Sequences for shRNAs Abbreviations: PKM2, pyruvate kinase isozyme type M2; shPKM2, shRNA against PKM2. |
  | Figure S1 PKM2 high expression is associated with poor survival in CC. Notes: Kaplan–Meier plots were generated using the web-based tool GEPIA (http://gepia.cancer-pku.cn/). The results revealed an association between high expression of PKM2 and unfavorable overall survival (A) and disease-free survival (B) in patients with CC. Abbreviations: CC, cervical cancer; PKM2, pyruvate kinase isozyme type M2. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.