Back to Journals » OncoTargets and Therapy » Volume 9
Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy
Authors Jiang P, Wang P, Sun X, Yuan Z, Zhan R, Ma X, Li W
Received 13 September 2015
Accepted for publication 22 December 2015
Published 13 June 2016 Volume 2016:9 Pages 3501—3509
DOI https://doi.org/10.2147/OTT.S96278
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Pengfei Jiang,1,* Ping Wang,2,* Xiaoling Sun,3 Zhongshun Yuan,4 Rucai Zhan,5 Xiangyu Ma,2 Weiguo Li2
1Medical Department, Yuhuangding Hospital, Yantai, 2Neurosurgery Department, Qilu Hospital, Shandong University, Jinan, 3Neurosurgery Department, Yuhuangding Hospital, Yantai, 4Neurosurgery Department, Yinan People’s Hospital, Linyi, 5Neurosurgery Department, No 3 Hospital of Jinan, Jinan, People’s Republic of China
*These authors contributed equally to this work
Abstract: Temozolomide (TMZ) is commonly used in glioma chemotherapy. However, a great clinical challenge for TMZ is chemoresistance. H19 transcripts are recognized as long noncoding RNAs, which potentially interact with chromatin-modifying complexes to regulate gene expression via epigenetic changes. Our data based on glioma patients showed that the expression of H19 was significantly upregulated in TMZ-resistant tumors compared with the TMZ-sensitive tumors. To determine the function of H19 in glioma, cell lines U87 and U251 were exposed to TMZ to establish TMZ-resistant clones U87TMZ and U251TMZ. In U87TMZ and U251TMZ, the expression level of H19 transcripts was increased compared to wild-type or nonresistant clones, as determined by real-time quantitative reverse transcription polymerase chain reaction. Concomitant treatment with small interfering RNA specifically targeting H19 and TMZ in resistant glioma clones resulted in decreased IC50 values for TMZ, and increased apoptotic rates than control small interfering RNA-treated cells. This was also evident by the increased PARP cleavage in resistant cells exposed to TMZ + si-H19. Furthermore, the reduced expression of H19 altered major drug resistance genes, such as MDR, MRP, and ABCG2, both at the mRNA and protein levels. Taken together, these findings suggest that H19 plays an important role in the development of TMZ resistance, and may represent a novel therapeutic target for TMZ-resistant gliomas.
Keywords: glioma, temozolomide, lncRNA, H19, drug resistance
Introduction
Glioma is the most common type of primary brain tumor, and glioblastoma multiforme (GBM) is the most aggressive form with very poor survival rates.1,2 Patients diagnosed with glioma are confronted with surgical resection, concurrent irradiation, and adjuvant temozolomide (TMZ) as the treatment protocol. TMZ induces DNA strand breaks in growing tumor cells, which goes unrepaired due to the O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, giving hope to many patients undergoing glioma treatment.3 However, overexpression of MGMT is detected in 60% of glioblastomas, making them inherently resistant to chemotherapy including TMZ in this subset of population.4 Further, like in any other cancer, recurrence due to resistance also impacts the therapeutic efficacy of TMZ in GBM. Hence, better understanding of the mechanisms related to chemoresistance is warranted to evade drug resistance mechanisms, and possibly find a cure.
Long noncoding RNA (LncRNA) is defined as a class of non-protein-coding transcripts >200 nucleotides long. An increasing number of lncRNAs have been identified to be involved in some important biological processes, such as imprinting control, cell differentiation, development, and tumorigenesis.5 Apart from this, lncRNAs have multifunctional roles acting as mediators in cellular signaling, molecular decoys, molecular guides at the chromatin sites, and during scaffold modeling.6 Alterations of lncRNAs have been linked with several diseases including cancer.7 These have definite roles in the pathogenesis of cancerous as well as normal cell functions but with no functional protein-coding ability.8,9 Furthermore, some of the lncRNAs are of prognostic significance, indicating a promising role in the glioma tumorigenesis.10 H19 RNA has been characterized as an oncogenic lncRNA encoding 2.3 kb long transcript that does not contain any known open reading frames.11,12 H19 has riboregulatory and pro-tumorigenic roles in other cancers such as hepatocellular carcinoma, bladder, and breast cancers.13,14 The expression of H19 is highly upregulated in gliomas, as it can bind with transcription factor c-Myc to drive tumor transformation and contribute to tumorigenic phenotypes.15 Recent report indicates that H19 regulates glioma development with the help of a H19-derived miR-675, and depletion of H19 via small interfering RNA (siRNA) decreases invasion rates of glioma.16 Although this provides important clues for the understanding of H19-miRNA network, the function of H19 in the TMZ-induced resistance in glioma is largely unknown.
In the current study, we determined the expression level of H19 in TMZ-resistant patient samples, and explored the biological function and potential mechanism of drug resistance in glioma. We found that H19 showed aberrant expression in TMZ-resistant patients and cell lines. Specifically, silencing H19 by concomitant treatment with TMZ chemotherapy and siRNA sensitized glioma cells to apoptosis. Furthermore, expression of important drug resistance genes such as MDR, MRP, and ABCG2 was downregulated on knockdown of H19. Taken together, our studies highlight the novel connecting link between H19 and TMZ drug resistance, which could be targeted in glioma.
Materials and methods
Selection of patients
A total of 69 patients with glioma were enrolled in this study. All these patients received TMZ-based chemotherapy, and no obvious primary tumor shrinkage was identified as TMZ resistance. The 25 TMZ-resistant and the 44 TMZ-sensitive glioma tissues were harvested from patients who were sensitive or inert to 200 mg/m2 monotherapy of TMZ, respectively, while surgical resection was carried out at the Yantai Yuhuangding Hospital, Yantai, Shandong, People’s Republic of China, during 2009–2013. Tissues were banked after obtaining written informed consent under an approved protocol by the Institutional Research Ethics Board. RNA expression profiles of three H19 transcript variants were detected by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Cell culture
Human neuronal glioma cell lines U87 and U251 were purchased from the Cell Culture Center (Chinese Academy of Medical Sciences, Beijing, People’s Republic of China). The use of these two cell lines in this study was approved by the Institutional Research Ethics Board of Yuhuangding Hospital. U87 and U251 cell lines were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). The cells were maintained under standard cell culture conditions at 37°C and 5% CO2 in a humid environment. For the treatment in cell lines, stock solution of TMZ (Schering-Plough, Kenilworth, NJ, USA) was prepared by dissolving the drug in dimethyl sulfoxide (Sigma-Aldrich, St Louis, MO, USA). U87 and U251 cells were initially exposed to 10 μM TMZ for 2 weeks, and then the TMZ concentration was increased by approximately twofold for every two passages until it reached 640 μM (maximum concentration kills all resistant cells, and the established cells were induced at 200 μM TMZ) to establish the TMZ-resistant cell lines.17
RNA extraction and qRT-PCR
TRIzol Reagent (Life Technologies) was used to harvest the adherent cells according to the standard protocol. The QuantiTect Reverse Transcription Kit (Qiagen) was used for reverse transcription strictly following manufacturer’s instructions. Reverse transcription products were diluted 1:20 in nuclease-free water. qRT-PCRs were performed using the LightCycler 480 (Roche) with an initial 2-minute denaturation step at 95°C, 40 cycles of a 15-second denaturation step at 95°C, followed by a 60-second hybridization step at 60°C. Expression levels were calculated as 2−ΔΔCt values, normalized to GAPDH genes. The primers used are shown in Table 1.
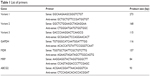  | Table 1 List of primers |
siRNA knockdown
siRNA oligonucleotides were purchased from Applied Biosystems with the Silencer® Select custom modification to enhance target specificity and recommended for noncoding RNA targets. Stocks were reconstituted in nuclease-free water to obtain 50 μM concentration and diluted to a 10 nM final concentration for all the transfections. For all experiments, control siRNA oligonucleotide transfections were performed using the same concentrations as the H19 siRNA oligonucleotides. The sequences of siRNAs targeting H19 are as follows (5′–3′): sense, GCGGGUCUGUUUCUUUACUUU; anti-sense, AGUAAAGAAACAGACCCGCUU. The sequences of negative control (NC) are as follows (5′–3′): sense, GCGUUCUGGUCUUACUGUUUU; anti-sense, AGAGAAUAAACCCGCAGACUU. Transfection or co-transfection of siRNA was performed according to the manufacturer’s protocol of lipofectamine 2000 (Invitrogen).
Western blotting
The concentration of cellular protein was detected by bovine serum albumin protein quantitation assay. Proteins obtained from cell lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies overnight after blocking with 2% bovine serum albumin, and thereafter probed for about an hour with secondary antibodies conjugated with horseradish peroxidase. Further, membranes were developed using chemiluminescence substrate, and imaged. All the primary antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA): PARP (sc-56196, mouse monoclonal antibody), MDR (sc-55510, mouse monoclonal antibody), MRP (sc-18835, mouse monoclonal antibody), and ABCG2 (sc-377176, mouse monoclonal antibody).
Detection of apoptosis by flow cytometry
Annexin V-PE apoptosis kit (BD Biosciences, San Jose, CA, USA) was used following manufacturer’s instructions. Briefly, U87TMZ and U251TMZ cells were transfected with siRNA-H19 (si-H19) or siRNA-Control (si-Con), respectively, and then treated with high-dose TMZ (640 μM) for 12 days prior to being harvested, and washed with cold PBS. The washed cells were resuspended in Annexin V binding buffer, followed by staining with Annexin V-PE simultaneously with 7-AAD. Further analysis was conducted using FACS (Becton-Dickinson, Franklin Lakes, NJ, USA), and the results were quantified using Cell Quest software (Becton-Dickinson).
MTT assay for cell cytotoxicity/viability
After transfection with H19 siRNA or control siRNA for 48 hours, cells (2×104 cells/well) were seeded in 96-well plates and incubated with or without TMZ for 24 hours at 37°C. Thereafter, 20 μL of 0.5 mg/mL MTT solution (Sigma-Aldrich) was added to each of the wells. The media was removed after 4 hours, and 0.2 mL DMSO was added to each well, and incubated for an additional 30 minutes. Absorbance was measured at 570 nm by a microplate reader (Bio-Rad, Hercules, CA, USA). The numerical values were used to construct the cell growth curve with cell viability. The numerical values = (OD value of test − OD value of blank)/(OD value of control − OD value of blank) were used to determine cell viability. Three replicate wells were set up in each group, and three independent experiments were performed repeatedly for the optimal statistical evaluation.
Statistical analysis
Statistical analysis was performed with SPSS software, version 17.0. The results are presented as mean ± standard deviation from three independent experiments, using Student’s t-test and/or one-way analysis of variance to perform appropriate data comparisons. P<0.05 was considered as statistically significant.
Results
H19 expression correlates with TMZ resistance in glioma patients
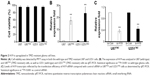
Initially, we analyzed the sequence of H19, and we found that there were three transcript variants of H19 (http://www.ncbi.nlm.nih.gov/gene/283120). Through rVista 2.0: Identification of Evolutionary Conserved Transcript website (http://rvista.dcode.org/) analysis, we found that the variant 1 represents the longest transcript, variant 2 uses an alternate splice site in the 3′ region resulting in a shorter transcript than variant 1, and variant 3 differs in the 5′ terminal exon, resulting in a shorter transcript than variant 1 (Figure 1A). So, the expression of all the three H19 variants was detected in both TMZ-resistant and TMZ-sensitive tumor tissues as shown in Figure 1B. While there was only negligible difference in the variants 2 and 3, variant 1 showed significantly larger difference between the sensitive and the resistant datasets analyzed. Importantly, expression of H19 variant 1 was significantly higher in TMZ-resistant tumor tissues than in TMZ-sensitive tumor tissues (P<0.001). Further, levels of variants 2 and 3 were also not abundant (the value of GAPDH was ~13, while the values of variants 2 and 3 were both ~30), and so, our progressive experiments focused only on the H19 variant 1, and henceforth designated as H19.
Expression of H19 is upregulated in TMZ-resistant cell lines
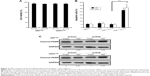
As a follow-up to our patient data that revealed a higher expression of H19 in the resistant population, we further assessed the expression of H19 in the TMZ-resistant glioma cell lines. U87 and U251 cell lines were constantly exposed to a high concentration of TMZ (200 μM) to establish the TMZ-resistant cell lines U87TMZ and U251TMZ. After the TMZ-resistant glioma cells were established, we tested the cell viability in both cell lines, and found that there was no significant change between them (Figure 2A). Interestingly, upregulated expression of H19 was also observed in U87TMZ and U251TMZ compared with wild-type U87 and U251 cells, respectively, via qRT-PCR assay (P<0.001 vs respective wild-type cells, Figure 2B). To further evaluate the function of H19 in TMZ-resistant glioma cells, we transiently transfected siRNA targeting H19 (si-H19) in U87TMZ and U251TMZ; the results showed that the resistant cells transfected with si-H19 showed remarkable downregulation of expression of H19 compared with cells transfected with control siRNA as assessed by qRT-PCR assay (P<0.001 vs respective control siRNA, Figure 2C).
Knockdown of H19 sensitizes TMZ-resistant clones to drug-mediated cytotoxicity
Given that H19 was overexpressed in TMZ-resistant patients and cell lines, we hypothesized that targeting H19 could sensitize U87TMZ and U251TMZ cells to TMZ-mediated cell cytotoxicity. Forty-eight hours after si-H19 transfection, cells were treated with the indicated concentration of TMZ, and the effects of H19 on the TMZ resistance of glioma cells were determined by MTT assay at 24 hours post-TMZ treatment, and the IC50 (half-maximal inhibitory concentration) was calculated by SPSS 17.0 software. As expected, si-H19 transfection in U87TMZ and U251TMZ cells rendered both cell lines more sensitive to TMZ-mediated cytotoxicity, as evidenced by the comparison of IC50 values between si-NC and si-H19-treated cells (Table 2).
Exposure of TMZ combined with H19 siRNA results in increased apoptosis
To decipher the biological impact of the function of H19 in TMZ resistance in glioma, additionally, we examined whether knockdown of H19 and TMZ exposure could lead to augmented apoptosis. We firstly confirmed that there was no significant cell viability change between the si-Con and si-H19 cells ( P>0.05, Figure 3A). We next treated the TMZ-resistant cells transfected with or without si-H19 at the dosage of 500 μM of TMZ, which is close to the IC50 of both cell lines. As shown in Figure 3B, the percent apoptosis of the cells was increased on concomitant treatment with si-H19 and TMZ in U87TMZ and U251TMZ cells, compared with control siRNA-transfected cells. Specifically, apoptotic rates were increased approximately eight- to nine fold just by knocking down H19 in both the resistant clones (4.1%±0.6% to 33.5%±3.6%, U87TMZ clone; 4.2%±1.2% to 38.5%±4.7%, U251TMZ clone; P<0.0001 vs respective control siRNA treatment). Corresponding with flow cytometry data, Western blotting also showed enhanced PARP cleavage after co-treatment with si-H19 and TMZ (Figure 3C). Individual TMZ or control siRNA treatment elicited only minor cleavage of PARP as indicated in the representative blots. Collectively, these results indicate that targeting H19 provides a sure shot strategy for sensitizing glioma cells to TMZ-mediated apoptosis.
Regulation of H19-induced TMZ resistance is partially mediated by major drug-resistant genes/proteins
It is reported that MDR, MRP, and ABCG2 genes are directly involved in the drug resistance mechanisms.18 Thus, we next explored whether any of these drug-resistant genes account for H19-mediated TMZ resistance. We transiently knocked down the expression of H19 in U87TMZ and U251TMZ cells, and the expression of MDR, MRP, and ABCG2 was determined via qRT-PCR and Western blots at the mRNA and protein levels, respectively. Our results suggested that knockdown of H19 significantly downregulated the expression of these drug-resistant genes, both at the mRNA (P<0.001 vs respective control siRNA, Figure 4A) and protein (Figure 4B) levels. These data confirm that the H19-induced TMZ resistance is in part mediated by MDR, MRP, and ABCG2.
Discussion
Malignant glioma is an incurable disease with very high infiltrative capacity exhibiting remarkable resistance to chemotherapy. The clinical progress is dismal and highly variable among patients,19 with ~32% of all diagnosed cases surviving only less than a year.20 This highly aggressive disease is often treated with a combination of chemotherapy, radiation, and surgery to improve the prognosis, and reduce recurrence and metastasis.21,22 TMZ is the frontline chemotherapy against GBM, and other difficult-to-treat glioma tumors.23 Few years back, a landmark study published results stating the benefits of TMZ, in addition to surgery and radiation, in a large cohort of patients.24 The existing standard of care for gliomas is limited to safe maximal surgical resection, followed with radiation, and concurrent and adjuvant TMZ.25 However, there still exists a paucity of certain clinical trials to determine the best possible therapy for the patient population diagnosed with glioma.26 Further, adding to the conundrum, recurrent gliomas contribute to the poor survival.27 And these are largely immune to TMZ because of developing chemoresistance in patients. Hence, strategies to neutralize and overcome chemoresistance require thorough understanding of the diverse concepts, and are a significant unmet need. While we know that the MGMT promoter methylation plays a crucial role in the drug resistance by removing alkyl groups from the O6 position of guanine, little is explored on other probable dependent mechanisms, which facilitate TMZ resistance to gliomas. Our current study explored the possibility of an lncRNA H19 to be involved in the chemoresistance of TMZ in glioma cells. Elevated expression of H19 was detected in a subset of TMZ-resistant glioma patients compared to TMZ-sensitive patients. This observation was fully recapitulated in the drug-resistant glioma cell line clones, which we developed. Treatment of H19 siRNA in combination with TMZ rendered glioma cells vulnerable to cytotoxicity and apoptotic cell death. Further, several important drug resistance genes were upregulated in the presence of H19, providing novel insights into the TMZ resistance in gliomas.
Emerging evidence indicate that lncRNA is one of the critical regulators in the progression of glioma. Ellis et al showed that lncRNA colorectal neoplasia differentially expressed (gene symbol: CRNDE) is one of the most upregulated lncRNA in gliomas, and other cancers such as colorectal carcinoma.28 It is activated and expressed in the specific regions within the human and mouse brain, although the mouse ortholog is the one which is largely distributed in induced pluripotent stem cells.28 Zhang et al reported that lncRNA HOX anti-sense intergenic RNA myeloid 1 (HOTAIRM1), highly expressed in the fetal brain, is closely associated with glioma grade, and increases with ascending glioma grade. Also, HOTAIRM1 reduction induced suppression of colony formation, G0/G1 phase cell cycle arrest, and orthotopic tumor growth inhibition.29 Zhen et al concluded that lncRNA nuclear enriched abundant transcript 1 (NEAT1) was upregulated in glioma tissues compared with noncancerous brain tissues. Additionally, knockdown of NEAT1 also reduced glioma cell proliferation, invasion, and migration.30 In GBM and normal brain tissue, one of the first reports suggested that lncRNAs have been differentially expressed, and it is believed that their regulatory roles are primarily involved in the pathogenesis of GBM.31 Yet another lncRNA H19 is a large intergenic noncoding RNA transcribed by RNA polymerase II, responsible for limiting placental growth before birth, by regulation of miR-675.32 Its main physiological function is unknown, but it has close functionality in the tumor initiation and fetal growth syndrome.6 H19 is believed to have both oncogenic and tumor suppressor roles; increased H19 expression is seen in gastric and bladder cancers, while negligible expression is associated with hepatocellular carcinoma.33–35 H19 primarily drives tumor transformation by binding with transcription factor c-Myc contributing to tumorigenesis.15 A recent microarray analysis comparing differential expression of lncRNAs in primary and recurrent glioma outlined several important lncRNAs that are critical for the glioma development. Surprisingly, H19 along with HOTAIRM1 and CRNDE was upregulated in glioma, and H19 was highly expressed in higher grade gliomas than lower grade gliomas.36 This study offers strong cues to the connecting link between glioma recurrence and expression of H19. However, the role of H19 in the TMZ-involved drug resistance has not been previously studied. Our studies revealed the crucial finding that H19 is highly expressed in TMZ-resistant tumors in comparison with TMZ-sensitive tumors in glioma patients. Of note, variant 1 was significantly altered compared with other variants, in glioma patients who were resistant to TMZ. Knockdown of H19 using specific siRNA in both U87 and U251 resistant cell lines led to decreased IC50 values for TMZ, indicating that H19 is the major determinant conferring oncogenic properties to the glioma cells. H19 is previously shown to have targeting sites for miR-17-92, and thus, H19 is indirectly involved in the differentiation, apoptosis, and proliferation of GBM neurospheres. Inhibition of miR-17-92/H19 effectively decreased cell viability, proliferation, and growth of neurospheres.37 Our data also go in line with another study claiming the oncogenic functions of H19 in breast cancer, when overexpressed. In particular, H19 mRNA-like noncoding RNA linked to E2F1 was responsible for the cell cycle progression seen in the breast cancer cells.13 In esophageal carcinoma cells, H19 induced epithelial–mesenchymal transition and provided impetus for the growth, invasion, and proliferation in vitro.38 A possible mechanistic pathway of H19 driving oncogenic signaling is through the suppression of RB, RUNX1, and CALN1 in gastric cancer.39,40 Thus, overexpression of H19 has been linked to anti-apoptotic and metastatic properties in gastric cancer.41 This is in part consistent with our observation in glioma cells, where apoptotic rates were dramatically increased in the presence of concomitant si-H19 and TMZ treatment.
One of the major obstacles of chemotherapy is the drug resistance phenomenon, assisted by MDR1 gene which encodes P-glycoprotein (P-gp), belonging to ATP-binding cassette drug efflux pump family.42 Recently, another class of multidrug resistance proteins was discovered, which are very versatile in transporting conjugated drugs, nucleotide analogs apart from P-gp-transported neutral organic compounds.43 In our study, major drug resistance genes such as MDR, MRP, and ABCG2 were downregulated in the TMZ-resistant glioma cell lines treated with H19 siRNA. Previously, H19 overexpression was suggested to possibly elicit MDR phenotype associated with p95 overexpression, deciphering for the first time a connecting link with multidrug resistance. In human breast MCF7/AdrVp, and lung NCI-H1688 cancer cell lines, overexpressing p95 but no demonstrable P-gp or MRP, H19 expression was found to be upregulated.44 Another study revealed that H19 knockdown suppressed MDR1/P-gp, and sensitized doxorubicin-resistant R-HepG2 cells to cytotoxicity. This clarified the role of H19 in the regulation of P-gp expression in HepG2 cells.45 But, so far to the best of our knowledge, this is the first study connecting TMZ drug resistance and H19 in glioma. Silencing H19 expression brought about downregulation of MDR genes in TMZ-resistant cell populations, reinstating the pivotal role of H19 in the drug resistance phenomenon observed. Although detailed mechanisms involved in the H19 upregulation and induction of TMZ resistance are largely unknown at this moment, our studies provide necessary insights for the future work to better understand the potential of targeting H19 in glioma.
Acknowledgment
This work was supported by grants from the Shandong Province Science and Technology Department (BS2011YY010).
Disclosure
The authors report no conflicts of interest in this work.
References
Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. Journal of Neuro-oncology. 2012;107(2):359–364. | ||
Tate MC, Aghi MK. Biology of angiogenesis and invasion in glioma. Neurotherapeutics: The Journal of the American Society for Experimental NeuroTherapeutics. 2009;6(3):447–457. | ||
Gerson SL. Clinical relevance of MGMT in the treatment of cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2002;20(9):2388–2399. | ||
Taylor JW, Schiff D. Treatment considerations for MGMT-unmethylated glioblastoma. Current Neurology and Neuroscience Reports. 2015;15(1):507. | ||
Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochimica et Biophysica Acta. 2015;1859(1):128–138. | ||
Park JY, Lee JE, Park JB, Yoo H, Lee SH, Kim JH. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Research and Treatment. 2014;2(1):1–6. | ||
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discovery. 2011;1(5):391–407. | ||
Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science (New York, N.Y.). 2008;319(5871):1787–1789. | ||
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews. Genetics. 2009;10(3):155–159. | ||
Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiology of Disease. 2012;48(1):1–8. | ||
Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology. 1990;10(1):28–36. | ||
Ariel I, Ayesh S, Perlman EJ, et al. The product of the imprinted H19 gene is an oncofetal RNA. Molecular Pathology: MP. 1997;50(1):34–44. | ||
Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. The Journal of Biological Chemistry. 2005;280(33):29625–29636. | ||
Matouk IJ, DeGroot N, Mezan S, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2(9):e845. | ||
Barsyte-Lovejoy D, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Research. 2006;66(10):5330–5337. | ||
Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9(1):e86295. | ||
Munoz JL, Rodriguez-Cruz V, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Temozolomide resistance in glioblastoma cells occurs partly through epidermal growth factor receptor-mediated induction of connexin 43. Cell Death & Disease. 2014;5:e1145. | ||
Choudhuri S, Klaassen CD. Structure, function, expression, genomic organization, and single nucleotide polymorphisms of human ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP) efflux transporters. International Journal of Toxicology. 2006;25(4):231–259. | ||
Frosina G. Limited advances in therapy of glioblastoma trigger re-consideration of research policy. Critical Reviews in Oncology/Hematology. 2015;96(2):257–261. | ||
Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes & Development. 2015;29(12):1203–1217. | ||
Burgio E, Migliore L. Towards a systemic paradigm in carcinogenesis: linking epigenetics and genetics. Molecular Biology Reports. 2015;42(4):777–790. | ||
Abhinav K, Yeh FC, Mansouri A, Zadeh G, Fernandez-Miranda JC. High-definition fiber tractography for the evaluation of perilesional white matter tracts in high-grade glioma surgery. Neuro-oncology. 2015;17(9):1199–1209. | ||
Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2000;6(7):2585–2597. | ||
Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England Journal of Medicine. 2005;352(10):987–996. | ||
Nagasawa DT, Chow F, Yew A, Kim W, Cremer N, Yang I. Temozolomide and other potential agents for the treatment of glioblastoma multiforme. Neurosurgery Clinics of North America. 2012;23(2):307–322, ix. | ||
Chamberlain MC. Treatment of newly diagnosed malignant glioma in the elderly people: new trials that impact therapy. International Journal of Clinical Practice. 2013;67(12):1225–1227. | ||
Haar CP, Hebbar P, Wallace GCt, et al. Drug resistance in glioblastoma: a mini review. Neurochemical Research. 2012;37(6):1192–1200. | ||
Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in CanceR, Neurobiology, and DEvelopment. Frontiers in Genetics. 2012;3:270. | ||
Zhang JX, Han L, Bao ZS, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-oncology. 2013;15(12):1595–1603. | ||
Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumor Biology. 2016;37(1):673–683. | ||
Han L, Zhang K, Shi Z, et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. International Journal of Oncology. 2012;40(6):2004–2012. | ||
Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biology. 2012;14(7):659–665. | ||
Iizuka N, Oka M, Tamesa T, Hamamoto Y, Yamada-Okabe H. Imbalance in expression levels of insulin-like growth factor 2 and H19 transcripts linked to progression of hepatocellular carcinoma. Anticancer Research. 2004;24(6):4085–4089. | ||
Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Letters. 2013;333(2):213–221. | ||
Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. Journal of Translational Medicine. 2013;11:225. | ||
Chen Y, Wu JJ, Lin XB, et al. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. International Journal of Clinical and Experimental Medicine. 2015;8(4):5033–5043. | ||
Ernst A, Campos B, Meier J, et al. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29(23):3411–3422. | ||
Huang C, Cao L, Qiu L, et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncology Letters. 2015;10(1):291–296. | ||
Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. | ||
Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochemical and Biophysical Research Communications. 2014;448(3):315–322. | ||
Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle (Georgetown, Tex.). 2015;14(19):3112–3123. | ||
Bradley G, Juranka PF, Ling V. Mechanism of multidrug resistance. Biochimica et Biophysica Acta. 1988;948(1):87–128. | ||
Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. Journal of the National Cancer Institute. 2000;92(16):1295–1302. | ||
Ross DD, Gao Y, Yang W, Leszyk J, Shively J, Doyle LA. The 95-kilodalton membrane glycoprotein overexpressed in novel multidrug-resistant breast cancer cells is NCA, the nonspecific cross-reacting antigen of carcinoembryonic antigen. Cancer Research. 1997;57(24):5460–5464. | ||
Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26(33):4877–4881. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.