Back to Journals » OncoTargets and Therapy » Volume 13
Knockdown of Circular RNA Hsa_circ_0000714 Can Regulate RAB17 by Sponging miR-370-3p to Reduce Paclitaxel Resistance of Ovarian Cancer Through CDK6/RB Pathway
Authors Guo M, Li S, Zhao X, Yuan Y, Zhang B, Guan Y
Received 3 October 2020
Accepted for publication 8 December 2020
Published 24 December 2020 Volume 2020:13 Pages 13211—13224
DOI https://doi.org/10.2147/OTT.S285153
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Min Guo,1 Shuo Li,1 Xiaoyun Zhao,2 Ying Yuan,1 Bin Zhang,1 Yifu Guan1
1Department of Biochemistry and Molecular Biology, China Medical University, Shenyang, Liaoning 110122, People’s Republic of China; 2Department of Microbiology and Cell Biology, Shenyang Pharmaceutical University, Shenyang, Liaoning 110016, People’s Republic of China
Correspondence: Yifu Guan
Department of Biochemistry and Molecular Biology, China Medical University, Shenyang, Liaoning 110122, People’s Republic of China
Tel +86-133-8688-1690
Email [email protected]
Purpose: Paclitaxel resistance in ovarian cancer has become an urgent clinical problem. This study investigated the regulatory effects of RAB17 on the non-coding RNA network of the paclitaxel-resistant ovarian cancer cell A2780/PTX.
Methods: Microarray analysis was used to identify differentially expressed genes in paclitaxel-resistant cell A2780/PTX compared to the parent paclitaxel-sensitive cell A2780. Quantitative real-time PCR and Western blot were used to measure the expression of related mRNAs and proteins. The CCK8 assay was used to determine cell survival ratios and drug resistance indices in ovarian cancer cells. The clone forming assay was used to analyze the cell clone proliferation. Flow cytometry was used to analyze the cell cycle. Dual-luciferase reporter gene assays evaluated the relationship between the genes.
Results: RAB17 is highly expressed in A2780/PTX cells. RAB17 knockdown increased the cell sensitivity to paclitaxel, inhibited proliferation, and caused cell cycle arrest in the G1 phase in A2780/PTX. Western blot confirmed that RAB17 influenced cell behavior by activating the CDK6/RB signaling pathway. Bioinformatics analyses identified RAB17 as a new target by the microRNA miR-370-3p, and the latter was predicted to interact with circular RNA hsa_circ_0000714. Hsa_circ_0000714 indeed acted as a miRNA sponge for miR-370-3p allowing its regulation of RAB17 expression. This regulation was accomplished through the CDK6/RB signaling pathway.
Conclusion: Hsa_circ_0000714 acts as a sponge for miR-370-3p, and regulates RAB17 expression through the CDK6/RB signaling pathway, which plays a role in the malignant progression of the paclitaxel-resistant ovarian cancer cell A2780/PTX.
Keywords: hsa_circ_0000714, miR-370-3p, RAB17, paclitaxel resistance, ovarian cancer
Introduction
Ovarian cancer is the seventh most common cancer affecting women worldwide.1 More than 220,000 new cases and approximately 152,000 deaths due to ovarian cancer have been reported each year.2,3 Paclitaxel, as the first-line therapeutic drug for ovarian cancer treatment, has shown remarkable efficacy. However, about 80% of ovarian cancer patients have developed paclitaxel resistance or even multiple-drug resistance after years of treatment,2,4 leading to treatment failure. Thus there is an urgent clinical problem that needs to overcome the paclitaxel resistance phenomenon in the clinical treatment of ovarian cancer.5
Our microarray results showed that RAB family proteins were significantly overexpressed in the paclitaxel-resistant ovarian cancer cell A2780/PTX compared with the paclitaxel-sensitive ovarian cancer cell A2780, and the most significant differences were found in RAB17 expression. RAB GTPases are composed of ~200 amino acids and their molecular weight is between 20,000–30,000 Daltons.6,7 They are nucleoside-dependent molecular switches that act as GTP binding (active) and GDP binding (inactive) states.8 Over 66 RAB proteins in humans have been described.9 RAB25 overexpression promoted the proliferation of ovarian cancer cells,10 whereas RAB21 downregulation was associated with the increased malignancy of human ovarian cancer.11 RAB17 is highly expressed in the liver, kidney, and intestine,12 and recent studies have suggested that it may be associated with the occurrence and development of a variety of human malignancies. To date, the biological functions of RAB17 and its potential role in paclitaxel resistance of ovarian cancer have not been reported.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs. Given their many characteristics, such as preservation, stability, and tissue-specific expression,13 they have attracted great interest for their application to clinical treatment and pharmaceutic development.14 CircRNAs have been described as competitive endogenous RNAs (ceRNAs) and interact with microRNAs (miRNAs) to negatively regulate their activity.15 MiRNAs, another class of non-coding RNAs of 22–25 nucleotides long, bind to the 3ʹ-untranslated regional (3ʹ-UTR) of target mRNAs through imperfect base pairing to regulate gene expressions.16 Thus, circRNAs, miRNAs, and target gene mRNAs could form regulatory axes for different biological functions, including cancer development. Previous studies have shown that the circRNAs/miRNAs/mRNAs cross-talk has a potential role as a tumor inhibitor and activator in ovarian cancer cell lines.17 And performed a systematic analysis of circRNAs expression in clinical ovarian tumor samples.18 In particular, circ_EPSTI1 was overexpressed in 50 ovarian cancer samples and was shown to sponge miR-942, which is downregulated in ovarian cancer tissue.19 Overexpression of circ_0051240 in ovarian cancer tissues promotes tumor formation, cell growth, progression, and invasion, this circRNA was able to sponge miR-637, which inhibited the expression of KLK4.20
In this study, we investigated RAB17 expression in paclitaxel-resistant as well as paclitaxel-sensitive ovarian cancer cells. Furthermore, the binding sites of miR-370-3p targeting the 3ʹ-UTR of RAB17 mRNA were identified. We also confirmed that hsa_circ_0000714 was highly expressed in the paclitaxel-resistant ovarian cancer cell, and could serve as a sponge of miR-370-3p to regulate the expression of RAB17, leading to the formation of the hsa_circ_0000714/miR-370-3p/RAB17 regulatory axis. We show that this axis promotes paclitaxel resistance in the ovarian cancer cell via activation of the CDK6/RB signaling pathway. Experimental results and conclusions in this study may contribute to a better understanding of the paclitaxel resistance mechanism of ovarian cancer and the development of new strategies for the paclitaxel-resistant ovarian cancer patients.
Materials and Methods
Cell Lines and Cell Culture
Two human epithelial ovarian cancer cell lines SKOV3 and A2780 were commercially acquired from Beijing Shijitan Hospital (Beijing, China) and the use of the cell lines was approved by the Medical Ethics Review Committee of China Medical University. Cells were cultured in RPMI-1640 and DMEM medium (Hyclone, Logan, UT, USA), and supplemented included with L-glutamine and 10% fetal bovine serum (TBD, Tianjin, China). The paclitaxel-resistant ovarian cancer cell lines SKOV3/PTX and A2780/PTX were prepared following a standard stepwise selection procedure. All cell lines were cultured in a humidified incubator with 5% CO2 at 37 °C. Cell lines grew in a monolayer and passage was performed when cultures were 70–80% confluent. The cell lines were tested for mycoplasma before use.
Establishment of Paclitaxel-Resistant Cells
A2780 cells were maintained in DMEM medium containing paclitaxel at a concentration of 0.1 nM for 24 h, paclitaxel was supplied by the Shenyang Sinopharm Group (Shenyang, China). The surviving cells were selected for the next survival selection step using a higher paclitaxel concentration. This selection process was repeated for several steps with an increment of 0.5 nM at each step until all cells could survive at the paclitaxel concentration of 500 nM. The surviving cells were able to maintain the paclitaxel-resistant phenotype in the absence of the selection pressure and were named as A2780/PTX. The same procedure was used to prepare SKOV3/PTX.
Cell Treatment
Lentiviruses carrying RAB17 expression vectors were obtained from GeneChem (Shanghai, China). RAB17 overexpression was conducted according to the manufacture’s instructions. Briefly, cells (0.5×105 cells/well) were seeded in a 12-well plate and treated with lentiviral particles to establish RAB17 overexpression [40 µL polybrene and 2.5 µL/well containing 1×108 infectious units (IFU) of RAB17 overexpression virus] and the negative control (40 µL polybrene and 2.5 µL/well containing 1×108 IFU negative virus) groups. The blank group consisted of A2780 cell with no exposure to lentiviruses transfection. Fresh medium DMEM was replaced in each infected well following 24 h of incubation. Stable cell lines were obtained by treatment with 5 μg/mL puromycin (Beyotime, Shanghai, China).
Cell Transfection
RAB17 siRNA (si-RAB17), negative control siRNA (si-NC), hsa_circ_0000714 siRNA (si-circ_0000714), negative control siRNA (si-NC) was obtained from GenePharma (Shanghai, China). MicroRNA-370-3p mimics and mimics negative control (mimics-NC) were purchased from RiboBio (Guangzhou, China). The primers used are listed in Table 1. A2780 and A2780/PTX cells were cultured in 6-well plates with serum-free medium overnight and then transfected with si-RAB17 (50 nM) or si-control (50 nM), si-circ_0000714 (50 nM) or si-control (50 nM), and miR-370-3p mimics (70 nM) or mimics-negative control (70 nM) for 48 h using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After at least 5 h of incubation, the medium was changed and cells were subsequently cultured at 37 °C for 48 h.Quantitative real-time polymerase chain reaction (qRT-PCR) was used to determine the transfection efficiency.
 |
Table 1 Primer Sequences Used for siRNA |
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using the Trizol method (Invitrogen), and reverse transcribed to cDNA using PrimeScript RT Master Mix (Takara, Dalian, China). Quantitative PCR was performed using TB Green® Premix Ex Taq™ II (TliRNaseH Plus) (Takara). The expression of RAB17, hsa_circ_0000714, hsa_circ_0001298, hsa_circ_0000540, hsa_circ_0000847, hsa_circ_0069905, hsa_circ_0000417 and miR-370-3p were determined by RT-PCR analyses using TB Green® Premix Ex Taq™ II (TliRNaseH Plus) (Takara). Mirna was reversed transcribed and amplified by Mir-X miRNA First-Strand Synthesis Kit and TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara), the primers for mRNA, miRNA, and circRNAs are listed in Table 2. U6 small nuclear RNA served as an internal control for miRNA expression detection, and GAPDH was used as an internal control for mRNA gene expression. The PCR process was performed on the LightCycler480 system (Roche, Basel, Switzerland). The PCR cycling was as follows: 95°C for 30 sec; followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec; and 1 cycle of 95°C for 5 sec, 60°C for 60 sec; lastly 1 cycle of 50°C for 30 sec. The relative RNA expression level was calculated with 2−∆∆ct.
 |
Table 2 Primer Sequences Used for qRT-PCR |
Cell Counting Kit-8 (CCK-8) Assay
The Cell Counting Kit-8 (CCK8) (Beyotime) was used to determine cell proliferation. Human ovarian cancer cells (1×104/well) were plated on 96-well plates for 48 hours with different treatments, and 10 μL/well CCK8 solutions were added at a specified time. Finally, the absorbance at 450 nm was measured after 4 h incubation.
Western Blot Analysis
Cells were harvested and washed with PBS. Protein was extracted following cell lysis using RIPA buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate) (Beyotime) containing protein inhibitors followed by ultracentrifuge at 13,000 rpm for 5 min. Supernatants were collected, and protein was quantified by the BCA assay (Beyotime). Samples were boiled at 100 °C for 5 min. The extracted proteins (30 µg per lane) were separated by SDS-PAGE electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were blotted with 5% non-fat milk at room temperature for 2 h. Primary antibodies targeting the proteins of interest were incubated with membranes with shaking at 4 °C overnight. Next, primary antibodies were collected, and membranes were incubated further with HRP tagged secondary antibodies at room temperature for an additional 2 h. The primary antibodies were diluted to 1:1000, secondary antibodies were diluted to 1:10,000. Finally, the immune reactive proteins were detected using an enhanced chemiluminescence kit (#WLA003a, Wanlei, Shenyang, China) and the enhanced chemiluminescence detection system (Tanon-5200, Tanon Science and Technology, Shanghai, China).
Western blotting was performed using the following antibodies: anti-RAB17 (#8013S, CST, Shanghai, China), anti-CDK6 (#ab124821, Abcam, Shanghai, China), anti-FOXO1A (#ab179450, Abcam), anti-Cyclin D1 (#ab40754, Abcam), anti-CDK4 (#ab108357, Abcam), anti-RB (#ab181616, Abcam), anti-p-RB (#ab184796, Abcam) and anti-GAPDH (#60004-1-lg, proteintech, Wuhan, China), anti-BCL-2 (#WL01556, Wanlei), anti-BCL-XL (#WL03353, Wanlei), anti-BAX (#WL01637, Wanlei), anti-N-cadherin (#WL01047, Wanlei), anti-E-cadherin (#WL01482, Wanlei). Secondary antibodies used in the experiments were from Santa Cruz (anti-rabbit, #ZB-2301 or anti-mouse, #ZB-2305; ZSGB Bio, Beijing, China).
Cell Colony Formation Assay
A2780/PTX cell was transfected with RAB17 siRNA for 48 h, then harvested and washed with PBS. Cells were planted in 6-well plates at a density of 400 cells per well, and cultured in RPMI-1640 supplemented with 10% FBS. After 2 weeks, the cells underwent fixation with 10% paraformaldehyde at room temperature for 30 min and stained with 0.25% of crystal violet at room temperature for 30 min. Finally, the newly formed colonies were imaged and counted. Using ImageJ software (version 1.46r, Bethesda, MD, USA), respectively, A2780/PTX cells were also transfected with si-negative control for comparison. The same experimental process applies to A2780.
Dual-Luciferase Reporter Gene Assay
The binding regions of RAB17 with miR-370-3p, hsa_circ_0000714 with miR-370-3p was predicted using the StarBase database (http://starbase.sysu.edu.cn/). The interactions between hsa_circ_0000714, miR-370-3p, and RAB17 were measured using the pMIR-REPORT system (GenePharma). The hsa_circ_0000714 and RAB17 mRNA 3′-UTR sequences containing wild-type (WT) or mutant-type (MUT), miR-370-3p binding sites were synthesized and respectively inserted into pMIR-report vectors. A2780 cells (1×105) were seeded into 96-wells and then co-transfected with miR-370-3p mimics or control mimics using Lipofectamine 2000 (Invitrogen). 48 h after transfection, the cells were harvested and lysed. Dual-luciferase activities were tested using a dual-luciferase reporter assay kit (Promega, Madison, Wisconsin, USA).
Flow Cytometry Assay
A total of 1×106 – 9×106 cells at the logarithmic phase of growth were harvested, rinsed twice with PBS, and fixed with 70% cold EtOH. The cells were then stained with 1 mL propidium iodide (50 µg/mL, KeyGen Bio, Jiangsu, China) in dark for 30 min. A FACS Calibur flow cytometer (Becton Dickinson and Company, New Jersey, USA) was employed to detect the cell cycle, which was then analyzed using MODFIT software (Becton Dickinson and Company).
Statistical Analyses
GraphPad (Prism Ver.8, GraphPad Prism Software, San Diego, CA, USA) was used to analyze statistical significance between groups. Experimental data were depicted as mean SD. Comparisons between two groups were analyzed by independent-samples t-test with Welch’s correction. And a p-value less than *p<0.05, **p<0.01 was considered to be statistically significant.
Results
RAB17 Was Highly Expressed in Paclitaxel-Resistant Ovarian Cancer Cells
First, we characterized the paclitaxel-sensitive ovarian cancer cells SKOV3 and A2780 as well as paclitaxel-resistant ovarian cancer cells SKOV3/PTX and A2780/PTX. The paclitaxel IC50 values of SKOV3 and SKOV3/PTX were determined using CCK8 assay to be 9.02±0.93 nM and 472.20±2.14 nM, indicating that the paclitaxel resistance index (RI) was 52.35. The paclitaxel IC50 values of A2780 and A2780/PTX were 11.51±0.86 nM and 1701±1.89 nM, respectively, corresponding to a paclitaxel RI of 147.78 (Figure 1A).
Microarray data showed that the expression profile of RAB family proteins in A2780 and A2780/PTX were dramatically different and that RAB17 high expression ranked first (Table 3). QRT-PCR and Western blotting confirmed that the expressions of RAB17 in the paclitaxel-resistant SKOV3/PTX and A2780/PTX cell lines were much higher than that in SKOV3 and A2780 cells (Figure 1B). Based on these results, we speculated that RAB17 might play an important role in the paclitaxel resistance of ovarian cancer.
 |
Table 3 The Up-Regulated and Down-Regulated Genes of A2780/PTX vs A2780 are Arranged According to the Ratio Change of the Microarray Data |
RAB17 Influenced Cell Paclitaxel Resistance Behavior, Colony Formation, and Cell Cycle in Ovarian Cancer Cells
A2780/PTX was infected with siRNA-RAB17 and A2780 was infected with lentivirus of RAB17 expression vectors. RAB17 expression was determined using qRT-PCR and Western blotting (Figure 2A). Si-RAB17 interference caused the paclitaxel IC50 value of A2780/PTX to decrease from 1716±2.40 nM to 640.6±2.56 nM, and the corresponding paclitaxel RI decreased from 147.78 to 55.66. In contrast, the paclitaxel IC50 value increased from 13.92±0.80 nM to 46.95±1.28 nM, when RAB17 was overexpressed of A2780 (Figure 2B). These results confirmed that RAB17 indeed influenced the paclitaxel resistance of ovarian cancer cells.
The colony formation assay provided results consistent with the aforementioned finding: si-RAB17 significantly inhibited the proliferation of A2780/PTX while RAB17 overexpression promoted the proliferation of A2780 cells (Figure 2C). Moreover, RAB17 knockdown of A2780/PTX led to cell cycle arrest: with fewer cells in the S phase and more in the G1 phase. In contrast, RAB17 overexpression of A2780 promoted the cell cycle with a reduction of the number of cells in the G1 phase and an increase in the S phase (Figure 2D). Furthermore, we observed that the expression of anti-apoptotic proteins was significantly decreased, and the expression of apoptotic proteins and adhesion proteins was increased in A2780/PTX following treatment with si-RAB17. Conversely, the expression of anti-apoptotic proteins increased, while the expression of apoptotic proteins and adhesion proteins decreased in A2780 cells following overexpression of RAB17 (Figure 2E). These results confirmed that RAB17 plays an important role in the development of paclitaxel resistance in ovarian cancer.
RAB17 Affected the Paclitaxel Resistance Behavior Through the CDK6/RB Signaling Pathway
To better understand the underlying mechanism of paclitaxel resistance induced by RAB17, we examined the expression of the paclitaxel resistance related protein P-gp. P-gp (also known as ABCB1, or MDR1) is a member of the superfamily of “classical” ABC transporters.21,22 This protein acts as a pump to efflux chemotherapeutic drugs out of cells and reduce their therapeutic efficacy.23 Western blotting results showed that in A2780/PTX cells exposed to si-RAB17 or when RAB17 was overexpressed in A2780 cells, the expression of P-gp changed according to the expression of RAB17, indicating that the paclitaxel resistance of A2780/PTX induced by RAB17 was related to P-gp levels (Figure 3A). However, our findings indicated that RAB17 had a stronger influence on the expression of cycle cell proteins than on P-gp protein, which encouraged us to further explore a potential novel mechanism involving RAB17 in paclitaxel resistance.
It is known that cyclin-dependent kinases (CDKs), a family of serine/threonine kinases, bind to the protein cyclins, and regulate the cell cycle during cancer progression. The CDK6-Cyclin D1 and CDK4-Cyclin D1 complexes play an important role in the G1 phase and G1/S transition of the cell cycle.24,25 Accordingly, we measured the expression of cell cycle related proteins: FOXO1A, CDK4, CDK6, Cyclin D1, RB, and the phosphorylated RB (p-RB S807).26
Our results showed that after A2780/PTX cell interference with si-RAB17, CDK6 expression was dramatically decreased, and that of its upstream protein FOXO1A as well as downstream proteins RB and p-RB were also decreased, together with the expression of Cyclin D1 involved in complex formation with CDK6. The expression of CDK4, however, was modestly affected compared with CDK6. The corresponding changes were also detected in the A2780 cell that overexpressed RAB17 (Figure 3B). Thus, we confirmed that RAB17 could affected paclitaxel resistance and various biological characteristics of ovarian cancer cells by activating the CDK6/RB signaling pathway.
RAB17 Was a New Target of miR-370-3p in Paclitaxel-Resistant Ovarian Cancer Cell
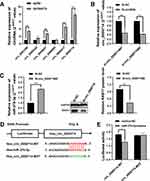
The StarBase database was searched to identify the potential miRNA binding regions targeting RAB17. The 3ʹ-UTR of RAB17 mRNA was found to bear potential binding sites for miR-370-3p (Figure 4A). To prove this hypothesis, we performed dual-luciferase assays using RAB17 reporters encoding the 3ʹ-UTR of RAB17. Our results showed that interference using miR-370-3p mimics significantly decreased luciferase activity of 3ʹ-UTR RAB17 wild-type (WT) reporter, which did not occur in when the reporter was exposed to the mimics negative control (NC), or when as the RAB17 reporter encoded a mutated 3ʹ-UTR sequence (Figure 4B). Overall, the results suggested that miR-370-3p might be a key miRNA regulator of RAB17 expression.
Upregulation of miR-370-3p Altered Cell Paclitaxel Resistance Behavior, Colony Formation and Cell Cycle of A2780/PTX
The inhibitory effect of miR-370-3p on tumor related genes has been demonstrated in a variety of cancer types including ovarian cancer.27–29 The effects of miR-370-3p on paclitaxel resistance in A2780/PTX were further examined. MiR-370-3p was highly expressed following exposure to miR-370-3p mimics, and accordingly, Western blotting showed the expression of RAB17 was significantly repressed (Figure 5A).
The CCK8 assay revealed that the paclitaxel IC50 value for A2780/PTX cells was reduced from 1653±1.86 nM to 828.5±0.88 nM, and the corresponding paclitaxel RI decreased from 147.78 to 71.98 (Figure 5B). Further, in the presence of with miR-370-3p mimics, the colony formation assay showed inhibition of A2780/PTX proliferation (Figure 5C), and flow cytometric data confirmed that the cells transfected A2780/PTX with miR-370-3p mimics led to cell cycle arrest and cell loss in the S phase, but increase in the G1 phase (Figure 5D). Western blotting revealed a reduced expression of FOXO1A, CDK6, Cyclin D1, RB, and other proteins, implying that the upregulation of miR-370-3p affected the CDK6/RB signaling pathway of A2780/PTX (Figure 5E).
Hsa_circ_0000714 Acted as a Potential Molecular Sponge of miR-370-3p
Searching the StarBase database identified six potential circRNAs predicted to interact with miR-370-3p, and qRT-PCR results confirmed that hsa_circ_0000714 was expressed in A2780/PTX most strikingly (Figure 6A).
Further, two siRNA oligonucleotides were prepared to target the unique back splicing junction of hsa_circ_0000714. The #2 antisense spliced junction siRNA decreased hsa_circ_0000714 expression more compared with the #1 antisense siRNA (Figure 6B). The knockdown of hsa_circ_0000714 in A2780/PTX decreased the expression of RAB17 at mRNA and protein levels (Figure 6C). These findings were consistent with those showing the significantly increased expression of miR-370-3p. To confirm molecular interaction between hsa_circ_0000714 and miR-370-3p, dual-luciferase reporter gene assay was performed, co-transfection of hsa_circ_0000714 wild-type (WT) with miR-370-3p mimics reduced strongly luciferase activity, however, no obvious effect on luciferase activity was found after co-transfection of hsa_circ_0000714 mutant-type (MUT) or the mimics negative control (Figure 6D and E). These results suggested that hsa_circ_0000714 could serve as a sponge of miR-370-3p and regulate RAB17 expression.
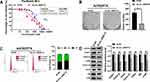
Hsa_circ_0000714 Influenced Paclitaxel Resistance Behavior, Colony Formation, Cell Cycle, and Signaling Pathway
To explore the role of hsa_circ_0000714 in ovarian cancer, we infected A2780/PTX cells with si-circ_0000714#2. CCK8 assays showed that the paclitaxel IC50 values decreased from 1799±2.13 nM to 802.8±2.72 nM when hsa_circ_0000714 was knocked down in A2780/PTX, the corresponding paclitaxel RI decreased from 147.78 to 69.75 (Figure 7A). Knockdown of hsa_circ_0000714 also significantly inhibited the proliferation ability of A2780/PTX (Figure 7B) and inhibited the cell cycle, blocking cells at the G1 phage (Figure 7C). Western blotting also demonstrated that knockdown of hsa_circ_0000714 decreased the expression of proteins of the CDK6/RB signaling pathway (Figure 7D).
Discussion
Paclitaxel is used as the first-line therapeutic agent for the treatment of ovarian cancer and has improved the overall response rate, clinical remission rate, and the median survival rate of ovarian cancer patients. Nonetheless, paclitaxel resistance may be developed after a period of clinical treatment.30,31 The mechanisms involved in paclitaxel resistance remains still unclear. It has been speculated that paclitaxel resistance could involve multiple pathways and multiple genes. The molecular mechanisms contributing to multiple-drug resistance include changes in the cell membrane and emergency response elements, changes of cell cycle checkpoint proteins, drug targets alternation, inhibition of cell apoptosis, increased repairing capability for DNA damage, and increased production of efflux protein ABC transporters, etc.32
Previous studies have shown that RAB17 plays an important role in the development of various cancers, but the results are controversial. In breast cancer cells, RAB17 is regulated by the ERK2 signaling cascade and acts as an inhibitor of cell motility.33 In hepatocellular cancer and non-small cell lung cancer, the downregulation of RAB17 promotes tumorigenesis and induces invasion, respectively.34,35 In the latest report, RAB17 was highly expressed in endometrial cancer, which regulated the cell cycle and promoted the proliferation of endometrial cancer by promoting the phosphorylation of AKT and mTOR.36 In our study, we observed a significantly higher expression of RAB17 in paclitaxel-resistant ovarian cancer cells A2780/PTX and SKOV3/PTX in comparison with paclitaxel-sensitive ovarian cancer cells A2780 and SKOV3 for the first time. We conducted a systematic investigation. To better understand the influence of RAB17 on paclitaxel resistance behavior. Cellular experiments determined that the higher expression of RAB17 was associated with a high paclitaxel resistance index, higher cell proliferation rate, and cell cycle alternation. Further experimental evidence suggested that RAB17 influenced cell cycle related proteins to create a paclitaxel-resistant phenotype in A2780/PTX cells via the CDK6/RB signaling pathway.
CircRNAs comprise a large class of non-coding RNAs.37 The emerging evidence on circRNA-microRNA-mRNA interactions provides new insights into the molecular pathways involved in the diagnosis, prognosis, and treatment of various cancers. In this study, using bioinformatics analysis combined with dual-luciferase experiments, we confirmed that the interaction of the 3ʹ-UTR of RAB17 mRNA with miR-370-3p regulates the paclitaxel resistance cellular behavior. MiR-370-3p has been studied intensively, and previous reports showed that it inhibits cell proliferation and induces the cell cycle arrest of human glioma cells.38 Further, miR-370-3p could inhibit the proliferation and invasion of thyroid cancer.39 In a recent report in ovarian cancer, miR-370-3p was sponged by hsa_circ_0061140, which inhibited the expression of FOXM1 and induced cell growth and metastasis.29 Furthermore, we observed circular RNA hsa_circ_0000714 was also significantly expressed in paclitaxel-resistant ovarian A2780/PTX cancer cells compared with A2780. The expression of hsa_circ_0000714 was negatively correlated with the expression of miR-370-3p, and the latter was also negatively correlated with the expression of RAB17. Thus, we propose the presence of a hsa_circ_0000714/miR-370-3p/RAB17 regulatory axis that could have an effect on paclitaxel resistance behavior via the CDK6/RB signaling pathway.
In summary, we observed that RBA17 is highly expressed in paclitaxel-resistant ovarian cancer cell A2780/PTX and that RAB17 affects the paclitaxel resistance behavior through the CDK6/RB signaling pathway. In paclitaxel-resistant ovarian cancer cell hsa_circ_0000714 can sponge miR-370-3p to further regulate RAB17, thus, forming the hsa_circ_0000714/miR-370-3p/RAB17 regulatory axis (Figure 8). To our knowledge, the findings and conclusions in the present study have not been reported previously. Our study may define a novel molecular mechanism for paclitaxel resistance and may provide the rationale for the development of new strategies for the clinical treatment of paclitaxel resistance in ovarian cancer. Further studies are needed to explore the complicated mechanism of the hsa_circ_0000714/miR-370-3p/RAB17 axis in vivo and to investigate its applications in ovarian cancer resistance, in resistance towards other anti-cancer agents, as well as other paclitaxel-resistant cancers.
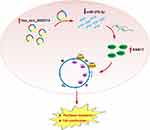 |
Figure 8 Schematic model illustrating the biological roles of the hsa_circ_0000714/miR-370-3p/RAB17 regulatory axis in A2780/PTX paclitaxel-resistant ovarian cancer cell. |
Acknowledgments
We thank the National Natural Science Foundation of China (Grant No. 31670821) and China Medical University (Grant No. QGZ2018076) for financial support.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3(7):502–516. doi:10.1038/nrc1123
3. Zhang XY, Zhang PY. Recent perspectives of epithelial ovarian carcinoma. Oncol Lett. 2016;12(5):3055–3058. doi:10.3892/ol.2016.5107
4. Sapiezynski J, Taratula O, Rodriguez-Rodriguez L, Minko T. Precision targeted therapy of ovarian cancer. J Control Release. 2016;243:250–268. doi:10.1016/j.jconrel.2016.10.014
5. Zahreddine H, Borden KL. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. 2013;4:28. doi:10.3389/fphar.2013.00028
6. Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128(17):3171–3176. doi:10.1242/jcs.166074
7. Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25(4):414–419. doi:10.1016/j.ceb.2013.04.002
8. Pylypenko O, Hammich H, Yu IM, Houdusse A. Rab GTPases and their interacting protein partners: structural insights into Rab functional diversity. Small GTPases. 2018;9(1–2):22–48. doi:10.1080/21541248.2017.1336191
9. Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 2012;10(1):71. doi:10.1186/1741-7007-10-71
10. Cheng KW, Lahad JP, Kuo WL, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10(11):1251–1256. doi:10.1038/nm1125
11. Hognas G, Tuomi S, Veltel S, et al. Cytokinesis failure due to derailed integrin traffic induces aneuploidy and oncogenic transformation in vitro and in vivo. Oncogene. 2012;31(31):3597–3606. doi:10.1038/onc.2011.527
12. Lehtonen S, Lehtonen E, Olkkonen VM. Vesicular transport and kidney development. Int J Dev Biol. 1999;43(5):425–433.
13. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
14. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi:10.1016/j.canlet.2016.12.006
15. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
16. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
17. Ahmed I, Karedath T, Andrews SS, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7(24):36366–36381. doi:10.18632/oncotarget.8917
18. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
19. Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi:10.1111/jcmm.14260
20. Obiezu CV, Scorilas A, Katsaros D, et al. Higher human kallikrein gene 4 (KLK4) expression indicates poor prognosis of ovarian cancer patients. Clin Cancer Res. 2001;7(8):2380–2386.
21. DeGorter MK, Xia CQ, Yang JJ, Kim RB. Drug transporters in drug efficacy and toxicity. Annu Rev Pharmacol Toxicol. 2012;52(1):249–273. doi:10.1146/annurev-pharmtox-010611-134529
22. Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455(1):152–162. doi:10.1016/0005-2736(76)90160-7
23. Kalabis GM, Kostaki A, Andrews MH, Petropoulos S, Gibb W, Matthews SG. Multidrug resistance phosphoglycoprotein (ABCB1) in the mouse placenta: fetal protection. Biol Reprod. 2005;73(4):591–597. doi:10.1095/biolreprod.105.042242
24. Clark AS, Karasic TB, DeMichele A, et al. Palbociclib (PD0332991)-a selective and potent cyclin-dependent kinase inhibitor: a review of pharmacodynamics and clinical development. JAMA Oncol. 2016;2(2):253–260. doi:10.1001/jamaoncol.2015.4701
25. Hamilton E, Infante JR. Targeting CDK4/6 in patients with cancer. Cancer Treat Rev. 2016;45:129–138. doi:10.1016/j.ctrv.2016.03.002
26. Chung M, Liu C, Yang HW, et al. Transient hysteresis in CDK4/6 activity underlies passage of the restriction point in G1. Mol Cell. 2019;76(4):562–573 e4. doi:10.1016/j.molcel.2019.08.020
27. Chen J, Liu G, Wu Y, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of beta-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18(1):150. doi:10.1186/s12943-019-1076-1
28. Song J, Kim D, Chun CH, Jin EJ. miR-370 and miR-373 regulate the pathogenesis of osteoarthritis by modulating one-carbon metabolism via SHMT-2 and MECP-2, respectively. Aging Cell. 2015;14(5):826–837. doi:10.1111/acel.12363
29. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi:10.1016/j.omtn.2018.08.010
30. Bowtell DD, Bohm S, Ahmed AA, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15(11):668–679. doi:10.1038/nrc4019
31. Bast RC, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. doi:10.1038/nrc2644
32. Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53(1):615–627. doi:10.1146/annurev.med.53.082901.103929
33. von Thun A, Birtwistle M, Kalna G, et al. ERK2 drives tumour cell migration in three-dimensional microenvironments by suppressing expression of Rab17 and liprin-beta2. J Cell Sci. 2012;125(6):1465–1477. doi:10.1242/jcs.092916
34. Qi J, Zhao P, Li F, et al. Down-regulation of Rab17 promotes tumourigenic properties of hepatocellular carcinoma cells via ERK pathway. Int J Clin Exp Pathol. 2015;8(5):4963–4971.
35. Wang M, Wang W, Ding J, Wang J, Zhang J. Downregulation of Rab17 promotes cell proliferation and invasion in non-small cell lung cancer through STAT3/HIF-1alpha/VEGF signaling. Thorac Cancer. 2020;11(2):379–388. doi:10.1111/1759-7714.13278
36. Zhou X, Xia G, Liu Y, et al. Analysis of carcinogenic signaling networks in endometrial cancer identifies RAB17 as a potential target. J Cell Physiol. 2020. doi:10.1002/jcp.29845
37. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
38. Peng Z, Wu T, Li Y, et al. MicroRNA-370-3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting beta-catenin. Brain Res. 2016;1644:53–61. doi:10.1016/j.brainres.2016.04.066
39. Chen F, Feng Z, Zhu J, et al. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol Ther. 2018;19(12):1139–1152. doi:10.1080/15384047.2018.1480888
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.