Back to Journals » Drug Design, Development and Therapy » Volume 16
Keloid Patient Plasma-Derived Exosomal hsa_circ_0020792 Promotes Normal Skin Fibroblasts Proliferation, Migration, and Fibrogenesis via Modulating miR-193a-5p and Activating TGF-β1/Smad2/3 Signaling
Authors Hu H, Mao G, Zheng J, Guo F
Received 18 August 2022
Accepted for publication 29 November 2022
Published 8 December 2022 Volume 2022:16 Pages 4223—4234
DOI https://doi.org/10.2147/DDDT.S386786
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Tin Wui Wong
Huan Hu,* Guangyu Mao,* Jianghong Zheng, Feng Guo
Department of Plastic Surgery, Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, 200233, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianghong Zheng; Feng Guo, Department of Plastic Surgery, Shanghai Sixth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, 600 Yishan Road, Xuhui District, Shanghai, 200233, People’s Republic of China, Email [email protected]; [email protected]
Background: Keloids are fibroproliferative disorders, which seriously affect the quality of life of patients with keloids. Additionally, circRNAs are enriched within exosomes derived from human blood samples, whereas their relationship with keloids remains largely unknown. It has been reported that hsa_circ_0020792 was abnormally upregulated in keloid tissues. However, the role of keloid patient plasma-derived exosomal hsa_circ_0020792 in the formation and development of keloids is not well understood.
Methods: Exosomes were isolated from the peripheral blood plasma of the patients with keloids (keloid patient-Exo) and healthy controls (Healthy control-Exo). The hsa_circ_0020792 and miR-193a-5p levels in keloid patient-Exo and healthy control-Exo, as well as in keloid fibroblasts and normal skin fibroblasts (NFs) were evaluated by RT-qPCR.
Results: The level of hsa_circ_0020792 was remarkably increased in keloid patient-Exo and keloid fibroblasts compared with that in Healthy control-Exo and NFs, respectively. In addition, keloid patient-Exo obviously enhanced the viability, migration, and extracellular matrix (ECM) synthesis, but reduced the apoptosis of NFs. Moreover, keloid patient-Exo notably promoted the fibrogenesis of NFs, as characterized by enhanced TGF-β signaling, increased expressions of phosphorylated Smad2/3. However, downregulation of hsa_circ_0020792 markedly reversed the promoting effects of keloid patient-Exo on cell growth, migration, and myofibroblast activation and fibrogenesis. Furthermore, downregulation of hsa_circ_0020792 significantly reduced the viability, migration, and fibrogenesis in NFs, whereas these phenomena were reversed by miR-193a-5p inhibitor.
Conclusion: Collectively, keloid patient plasma-derived exosomal hsa_circ_0020792 could promote the proliferation, migration, and fibrogenesis of NFs via modulating miR-193a-5p and activating TGF-β 1/Smad2/3 signaling.
Keywords: keloid, fibroblasts, exosomes, hsa_circ_0020792, miR-193a-5p
Introduction
Scarring in the skin after various trauma not only affects the external skin of the patient, but also seriously affects the quality of life of patients.1–3 In addition, pathological scar mainly includes keloid and hypertrophic scar, which is a spontaneous and excessive skin fibro-proliferative disease after trauma.4,5 Clinically, keloid is considered a “benign tumor” of the skin.5,6 The reason is that keloids grow beyond the boundary of the original wound, and there is no sign of spontaneous decay.7–9 Currently, the treatments for keloid include surgery and laser therapy; however, the therapeutic effects are not satisfactory.10,11
Circular RNAs (circRNAs) play important roles in mediating cell proliferation, migration, and invasion.12–14 CircRNAs can bind intracellular microRNAs (miRNAs) as competing endogenous RNAs (ceRNAs), thereby blocking the inhibition of miRNAs on their target genes.15 Moreover, it has been shown that circRNAs are related to the formation and development of keloid.16,17 For instant, circCOL5A1 could accelerate the development process of keloid by downregulating miR-7-5p.16 Besides, knockdown of circPDE7B could inhibit the progression of keloid by increasing miR-661.18
Exosomes are nano-sized membrane vesicles containing active substances (DNA, RNA, proteins, lipids, etc.).19,20 Additionally, exosomes are able to provide a new method for transferring effector information between cells in biological functions and normal body responses.21,22 For example, exosomal miRNA-21 can accelerate the proliferation of keloid fibroblasts by downregulating Smad7.23 Exosomal miR-29a derived from mesenchymal stem cells was able to suppress the excessive proliferation of human hypertrophic scar fibroblasts.24 Shi et al found that hsa_circ_0020792 is abnormally upregulated in keloid tissues.17 However, the role of keloid patient-derived exosomal hsa_circ_0020792 in the formation and development of keloid remains unclear. Thus, we aimed to explore whether keloid patient-derived exosomal hsa_circ_0020792 could regulate NFs proliferation, migration, and ECM production.
Materials and Methods
Samples
All specimens used in this study were obtained from Shanghai Sixth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine. The peripheral blood was collected from three patients with keloids (3 females, range 24–34 years old) and three healthy participants (3 females, range 25–32 years old). This study complies with the declaration of Helsinki and was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital. Informed consent was obtained from all participants.
Exosomes Extraction and Identification
The Exosome Isolation and Purification Kit (cat. no. UR52136-30T; Umibio, China) was applied to isolate exosomes from the peripheral blood plasma samples of patients with keloids and healthy participants (Keloid patient-Exo and Healthy control-Exo). Briefly, samples were centrifuged at 3000 x g for 10 min, and 10,000 x g for 10 min. Next, the supernatant was treated with Blood PureExo Solution at 4°C for 2 h, and then centrifuged at 10,000 x g for 1 h. Finally, the exosome vesicles were obtained and resuspended in PBS.
The isolated vesicles were suspended in 2% PFA and then added to a formvar-carbon copper grid. After that, the grid was treated with 1% glutaraldehyde for 5 min. Next, the grid was treated with uranium oxalate (pH = 7) for 5 min, and then treated with methylcellulose reagent for 10 min. Subsequently, a transmission electron microscopy (TEM) was used to observe the morphology of the isolated vesicles.
Next, the ZetaView analyzer was calibrated using polystyrene microspheres. Then, the vesicles were diluted by 1× PBS buffer. Subsequently, Nanoparticle Tracking Analysis (NTA) was used to observe the number and size of vesicles according to the previous report.25
Western Blot Assay
The protein concentration was quantified using the BCA protein assay kit (cat. no. C503021, Sangon biotech). Later on, 10% SDS-PAGE was used to separate protein. Next, protein was transferred onto the PVDF membranes. After that, the membranes were incubated with primary antibodies against CD9 (cat. no. 20597-1-AP, Proteintech), CD63 (cat. no. 67605-1-Ig, Proteintech), CD81 (cat. no. 66866-1-Ig, Proteintech), Collagen I (cat. no. ab260043, Abcam), Collagen III (cat. no. 22734-1-AP, Proteintech), α-SMA (cat. no. 23081-1-AP, Proteintech), TGF-β1 (cat. no. 21898-1-AP, Proteintech), p-Smad2 (cat. no. ab280888, Abcam), Smad2 (cat. no. 12570-1-AP, Proteintech), Smad3 (cat. no. 66516-1-Ig, Proteintech), p-Smad3 (cat. no. ab52903, Abcam), Bcl-2 (cat. no. 12789-1-AP, Proteintech), Bax (cat. no. 50599-2-lg, Proteintech), cleaved caspase 3 (cat. no. ab2302, Abcam), and GAPDH (cat. no. 60004-1-1, Proteintech) overnight at 4°C. The membranes were then immersed with an HRP-conjugated secondary antibody (A0216, Beyotime) at room temperature for 2 h. Afterwards, blot signals were observed by an ECL kit (cat. no. AS1059, ASPEN). β-actin was used as the internal standard. The western ladder (cat. no. WJ103, Shanghai Epizyme Biomedical Technology Co., Ltd) was used in this study.
Cell Culture
Fibroblasts collected from normal skin tissues were obtained from BeNa Culture Collection (China) and human keloid fibroblasts were obtained from Procell (China). Cells were cultured in DMEM (cat. no. L110, Shanghai BasalMedia Technologies Co., LTD) in 5% CO2 at 37°C. DMEM was contained 10% FBS (cat. no. 1803122, Biological Industries-Israel), 1% penicillin and 1% streptomycin (cat. no. 15070063, Gibco).
Cell Transfection
NFs was transfected with siRNA negative control (siR-NC), hsa_circ_0020792 siRNA1 (siR-circ-1), hsa_circ_0020792 siRNA2 (siR-circ-2) or hsa_circ_0020792 siRNA3 (siR-circ-3), miR-193a-5p inhibitor (miR-inhibitor) and miRNA negative control (miR-NC) using Lipofectamine 2000 (Thermo Fisher Scientific), respectively. The sequence as follows: siRNA-NC, ATTACAAGCGTTCACTCATTA; siR-circ-1, AAGCACAGCAGCATCTTCAAA; siR-circ-2, AAGCACTCATACTTTATGCAT; siR-circ-3, AAGCTCAGAAATTGGCTTTAA; miR-inhibitor, 5′-UCAUCUCGCCCGCAAAGACCCA-3′.
Reverse Transcription-Quantitative PCR (RT-qPCR)
The total RNA was extracted using Trizol reagent (cat. no. 15596026, Invitrogen). Next, the corresponding cDNAs were synthesized using the ReverTra Ace qPCR RT Kit (cat. no. FSQ-101, TOYOBO). Later on, qPCR was performed on the PCR System (ABI) using a SYBR Green PCR MASTER kit (cat. no. 4368708, Applied Biosystems). The primer sequence as follows: hsa_circ_0020792, forward, 5′-GGCCCGTGTTTGACTCAACT-3′ and reverse, 5′-CTGGGGAAGTTGTCGAAGATCA-3′; GAPDH, forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′; miR-193a-5p, forward, 5′-TATATGGGTCTTTGCGGGCG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6, forward, 5′-CTCGCTTCGGCAGCACAT-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 assay was applied to determine cell viability. NFs were seeded into 96-well plates (5×103 cells per well) overnight, and then treated with indicated exosomes (10 μg/mL) for 24 h. After that, cells were mixed with 10 μL of CCK-8 reagent (cat. no. MA0218-T, meilunbio®) for 2 h at 37°C. Next, the absorbance was detected at 450 nm with a microplate reader (Infinite M Nano, TECAN).
Flow Cytometry Assay
The Annexin V-FITC apoptosis detection kit (cat. no. C1062S, Beyotime) was used for evaluating cell apoptosis. NFs were incubated with indicated exosomes (10 μg/mL) for 24 h. After that, cells were stained with 5 μL annexin V-FITC and PI reagent for 15 min. Subsequently, a flow cytometer (BD Bioscience) was used to detect the apoptotic cells.
Transwell Migration Assay
Transwell assay was used for assessing cell migratory ability. The 24-well transwell chamber (cat. no. 3422, Corning) was used to perform transwell migration assay. NFs (1×105 cells per well) were plated onto the upper chamber and cultured with serum free medium. Meanwhile, 500 μL of medium with 10% FBS was loaded into the lower chamber. Later on, cells that had migrated from the upper part of the membrane to the lower part were stained with 0.1% crystal violet (cat. no. AS1086, ASPEN) at 24 h. Next, a microscope (ECLIPSE TS2, Nikon) was used to observe the stained migrated cells.
Statistical Analysis
The results data were analyzed using GraphPad Prism using One-way analysis of variance (ANOVA) and Tukey’s tests. Data were presented as mean ± SEM. *P < 0.05 was considered significant. All data were repeated in triplicate.
Results
Exosomes are Successfully Collected from Peripheral Blood of Healthy Participants and Patients with Keloid
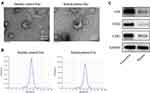
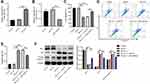
To explore the role of exosomes in the formation and development of keloid, exosomes were collected from the peripheral blood of the patients with keloid (keloid patient-Exo) and healthy controls (Healthy control-Exo). The results of TEM and NTA indicated that these isolated vesicles (50–150 nm in diameter) have lipid-bilayer membrane structures (Figure 1A and B). Meanwhile, these isolated vesicles were expressed exosome marker proteins CD9, CD63 and CD81 (Figure 1C). Collectively, exosomes were successfully isolated from peripheral blood of healthy participants and patients with keloid.
Hsa_circ_0020792 Level is Increased and miR-193a-5p Level is Reduced in Exosomes Derived from Peripheral Blood of the Patients with Keloid
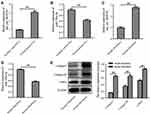
The data from Starbase (https://starbase.sysu.edu.cn) showed that there are two potential binding sites for miR-193a-5p in hsa_circ_0020792 (Supplementary Figure 1), suggesting that hsa_circ_0020792 might be a target of miR-193a-5p. Thus, we investigated whether keloid patient-Exo are enriched with hsa_circ_0020792, and whether keloid patient-Exo could regulate the development of keloids via targeting miR-193a-5p. As indicated in Figure 2A and B, hsa_circ_0020792 level was greatly elevated and miR-193a-5p level was obviously reduced in keloid patient-Exo compared to healthy control-Exo. In addition, hsa_circ_0020792 was increased and miR-193a-5p was decreased in keloid fibroblasts compared with that in NFs (Figure 2C and D). Moreover, the levels of ECM proteins Collagen I, Collagen III and α-SMA were elevated in keloid fibroblasts compared to NFs (Figure 2E). Furthermore, hsa_circ_0020792 level was significantly increased in TGF-β1-treated NFs (Supplementary Figure 2). To sum up, hsa_circ_0020792 level was increased and miR-193a-5p level was reduced in keloid patient-Exo and keloid fibroblasts.
Keloid Patient-Derived Exosomal hsa_circ_0020792 Enhances the Viability and Reduces the Apoptosis of NFs
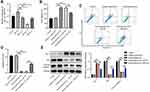
Next, to investigate whether hsa_circ_0020792-enriched keloid patient-Exo could affect the proliferation and collagen synthesis in NFs, NFs were transfected with siR-circ-1, siR-circ-2 or siR-circ-3. As shown in Figure 3A, siR-circ-2 remarkably reduced the level of hsa_circ_0020792 in NFs. Additionally, keloid patient-Exo obviously enhanced the viability and reduced the apoptosis of NFs compared to healthy control-Exo, whereas these phenomena were reversed by siR-circ-2 (Figure 3B–D). Meanwhile, keloid patient-Exo notably upregulated the level of Bcl-2 and downregulated the level of Bax and cleaved caspase 3 in NFs, whereas siR-circ-2 was able to reverse these changes (Figure 3E). All in all, keloid patient-derived exosomal hsa_circ_0020792 could promote the viability and inhibit the apoptosis of NFs.
Keloid Patient-Derived Exosomal hsa_circ_0020792 Promotes the Migration and Fibrogenesis of NFs via Activating TGF-β1/Smad2/3 Signaling
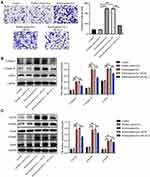
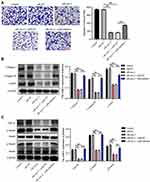
In order to explore the effect of keloid patient-derived exosomal hsa_circ_0020792 on the migration of NFs, transwell assay was performed. The results showed that keloid patient-Exo notably promoted the migration of NFs compared to healthy control-Exo (Figure 4A). However, siR-circ-2 abolished the pro-migratory effect of keloid patient-Exo on NFs (Figure 4A). In addition, compared to healthy control-Exo, keloid patient-Exo remarkably increased the levels of Collagen I, Collagen III, α-SMA, TGF-β1, p-Smad2 and p-Smad3 in NFs, whereas these increases were reduced by siR-circ-2 (Figure 4B and C). These data indicated that keloid patient-derived exosomal hsa_circ_0020792 could promote the migration and fibrogenesis of NFs via activating TGF-β1/Smad2/3 signaling.
Knockdown of hsa_circ_0020792 Inhibits the Viability, Migration, and Fibrogenesis of NFs via Upregulating miR-193a-5p
To clarify the effect of hsa_circ_0020792 and miR-193a-5p on the cell growth, apoptosis, migration and collagen synthesis on NFs, siR-NC or siR-circ-2 and miR-inhibitor or miR-NC were transfected into NFs. As illustrated in Figure 5A and B, siR-circ-2 significantly increased miR-193a-5p level in NFs, whereas miR-inhibitor obviously declined miR-193a-5p level in NFs. Besides, downregulation of hsa_circ_0020792 by siR-circ-2 notably reduced the viability and triggered the apoptosis of NFs, whereas these phenomena were abolished by miR-193a-5p inhibition (Figure 5C–E). Moreover, knockdown of hsa_circ_0020792 markedly downregulated the expressions of Bcl-2 and upregulated the expressions of Bax and cleaved caspase 3 in NFs, whereas these phenomena were reversed in the presence of miR-inhibitor (Figure 5F). Meanwhile, hsa_circ_0020792 knockdown notably suppressed the migration of NFs, whereas miR-inhibitor was able to reverse this effect (Figure 6A). Moreover, downregulation of hsa_circ_0020792 significantly reduced the level of Collagen I, Collagen III, α-SMA, TGF-β1, p-Smad2 and p-Smad3 in NFs, whereas these phenomena were reversed by miR-193a-5p inhibition (Figure 6B and C). Collectively, knockdown of hsa_circ_0020792 could reduce the viability, migration and fibrogenesis and induce the apoptosis of NFs via upregulating miR-193a-5p.
Knockdown of hsa_circ_0020792 Inhibits the Viability and Induces the Apoptosis of Keloid Fibroblasts via Upregulating miR-193a-5p
To further confirm the effect of hsa_circ_0020792 and miR-193a-5p on the growth of keloid fibroblasts, siR-circ-1, siR-circ-2 or siR-circ-3 was transfected into keloid fibroblasts. As revealed in Supplementary Figure 3A, siR-circ-2 sharply reduced hsa_circ_0020792 level in keloid fibroblasts. In addition, downregulation of hsa_circ_0020792 notably reduced the viability and induced the apoptosis of keloid fibroblasts, whereas these phenomena were reversed by miR-193a-5p inhibitor (Supplementary Figure 3B and C). To sum up, knockdown of hsa_circ_0020792 could inhibit the viability and induce the apoptosis of keloid fibroblasts via upregulating miR-193a-5p.
Discussion
It has been shown that peripheral circulating blood exosomes are involved in the pathogenesis of various diseases including skin diseases.26–28 CircRNAs have been found in exosomes derived from human blood samples with clinical implications.29 In addition, circRNAs exert key roles in the occurrence and development of keloid.30 Jiao et al reported that circCOL5A1 was able to suppress the proliferation, migration and ECM production of keloid fibroblasts through targeting miR-877-5p.31 Gao et al found that hsa_circ_0057452 could facilitate keloid progression by targeting miR-1225-3p.32 In addition, Shi et al found that hsa_circ_0020792 level was abnormally upregulated in the keloid tissues compared to healthy skin tissues.17 In this study, we found that hsa_circ_0020792 level was increased in exosomes derived from keloid patients compared to exosomes from healthy controls, which was consistent with the previous study. However, the role of exosomal hsa_circ_0020792 in the formation and development of keloids is not well understood.
The excessive ECM production and abnormal increase in fibroblast proliferation and migration are risk factors in the development of keloid.31,33 In addition, Xie et al found that keloid-derived fibroblasts expressed high levels of ECM proteins collagen I and collagen III,34 which was consistent with our results. Furthermore, we found that keloid patient-derived exosomal hsa_circ_0020792 obviously promoted the viability, migration and inhibited the apoptosis of NFs. Moreover, keloid patient-derived exosomal hsa_circ_0020792 notably accelerated the synthesis of ECM in NFs, as characterized by the increased levels of collagen I and collagen III. Meanwhile, α-SMA, a myofibroblast marker, exerts an important role in fibrogenesis.35 Our results showed that keloid patient-derived exosomal hsa_circ_0020792 obviously elevated α-SMA expression in NFs, suggesting that exosomal hsa_circ_0020792 promoted fibrogenesis in NFs. These results showed that hsa_circ_0020792-enriched exosomes derived from keloid patients were able to endow normal skin fibroblast cells with the biological properties of keloid fibroblasts.
Evidence has shown that TGF-β/Smads signaling plays vital roles in promoting the proliferation, collagen synthesis and fibrogenesis in keloid fibroblasts.36,37 Activation of TGF-β/Smads pathway could promote the keloid development.38 Chen et al showed that circ_0008450 knockdown could suppress the proliferation of human keratinized epithelial cells via inactivating the TGF-β/Smad2/3 signaling.39 In this study, we found that keloid patient-derived exosomal hsa_circ_0020792 remarkably upregulated the expressions of TGF-β1, p-Smad2 and p-Smad3 in NFs, whereas these changes were reduced by hsa_circ_0020792 knockdown. Collectively, hsa_circ_0020792-enriched exosomes derived from patients with keloids could promote the proliferation, migration, collagen synthesis and fibrogenesis of NFs via activating TGF-β1/Smad2/3 signaling.
It has been shown that miR-193a is associated with skin-related diseases.40,41 Polini et al showed that miR-193a-5p could exert a tumor suppressor role on cutaneous melanoma.42 However, the role of miR-193a-5p in keloid development remains unclear. In this study, our results showed that miR-193a-5p level was reduced in keloid patient-derived exosomes and keloid fibroblasts. Inhibition of miR-193a-5p was able to reverse the effect of hsa_circ_0020792 knockdown on the proliferation and migration of NFs, suggesting that downregulation of hsa_circ_0020792 could suppress the viability and migration of NFs via upregulating miR-193a-5p. Additionally, the inhibitory effects of hsa_circ_0020792 knockdown on the ECM synthesis and fibrogenesis of NFs were reversed by miR-193a-5p inhibitor, as shown by the increased levels of collagen I, collagen III, TGF-β1, p-Smad2 and p-Smad3, suggesting that hsa_circ_0020792 knockdown could suppress the ECM synthesis and fibrogenesis of NFs via upregulating miR-193a-5p. In pancreatic cancer, miR-193a was able to regulate tumor metastasis via targeting TGF-β2/SMAD2/3 signaling.43 Our results showed that key factors participated in TGF-β1/Smad2/3 pathway could be affected by hsa_circ_0020792 and miR-193a-5p. Furthermore, we further confirmed that hsa_circ_0020792 knockdown could inhibit the viability and induce the apoptosis of keloid fibroblasts via upregulating miR-193a-5p. These data showed that hsa_circ_0020792 could affect the progression of keloid via targeting miR-193a-5p.
Conclusion
In conclusion, keloid patient-derived exosomal hsa_circ_0020792 could promote normal skin fibroblasts proliferation, migration, and fibrogenesis via modulating miR-193a-5p and activating TGF-β1/Smad2/3 signaling. These results showed that hsa_circ_0020792 could affect keloid progression by miR-193a-5p/TGF-β1/Smad2/3 axis, suggesting a novel hsa_circ_0020792/miR-193a-5p/TGF-β1/Smad2/3 pathway underlying keloid treatment.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study complies with the declaration of Helsinki and was proved by Ethics Committee of Shanghai Sixth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (S2017-0023). The written informed consents have been obtained from patients and healthy donators.
Acknowledgments
Huan Hu and Guangyu Mao are co-first authors for this study.
Funding
This study was supported by National Natural Science Foundation of China (82172200).
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Li J, Chen L, Li Q, et al. Comparative peptidomic profile between human hypertrophic scar tissue and matched normal skin for identification of endogenous peptides involved in scar pathology. J Cell Physiol. 2018;233(8):5962–5971. doi:10.1002/jcp.26407
2. Meynköhn A, Fischer S, Neuss C, et al. Fractional ablative carbon dioxide laser treatment of facial scars: improvement of patients’ quality of life, scar quality, and cosmesis. J Cosmet Dermatol. 2021;20(7):2132–2140. doi:10.1111/jocd.13850
3. Oh H, Boo S. Quality of life and mediating role of patient scar assessment in burn patients. Burns. 2017;43(6):1212–1217. doi:10.1016/j.burns.2017.03.009
4. Cobbold CA. The role of nitric oxide in the formation of keloid and hypertrophic lesions. Med Hypotheses. 2001;57(4):497–502. doi:10.1054/mehy.2001.1373
5. Berman B, Maderal A, Raphael B. Keloids and hypertrophic scars: pathophysiology, classification, and treatment. Dermatol Surg. 2017;43(Suppl 1):S3–S18. doi:10.1097/DSS.0000000000000819
6. Liu X, Chen W, Zeng Q, et al. Single-cell RNA-sequencing reveals lineage-specific regulatory changes of fibroblasts and vascular endothelial cells in keloids. J Invest Dermatol. 2022;142(1):124–135.e111. doi:10.1016/j.jid.2021.06.010
7. Wang X, Liu K, Ruan M, Yang J, Gao Z. Gallic acid inhibits fibroblast growth and migration in keloids through the AKT/ERK signaling pathway. Acta Biochim Biophys Sin. 2018;50(11):1114–1120. doi:10.1093/abbs/gmy115
8. Berman B, Bieley HC. Keloids. J Am Acad Dermatol. 1995;33(1):117–123. doi:10.1016/0190-9622(95)90035-7
9. Scrimali L, Lomeo G, Nolfo C, et al. Treatment of hypertrophic scars and keloids with a fractional CO2 laser: a personal experience. J Cosmet Laser Ther. 2010;12(5):218–221. doi:10.3109/14764172.2010.514924
10. Yang Y, Jiang C, Xu Q. Combination therapy for bulky auricular keloids: a clinical experience. J Cosmet Laser Ther. 2019;21(1):14–16. doi:10.1080/14764172.2018.1439963
11. Sabry HH, Abdel Rahman SH, Hussein MS, Sanad RR, Abd El Azez TA. The efficacy of combining fractional carbon dioxide laser with verapamil hydrochloride or 5-fluorouracil in the treatment of hypertrophic scars and keloids: a clinical and immunohistochemical study. Dermatol Surg. 2019;45(4):536–546. doi:10.1097/DSS.0000000000001726
12. Yang G, Zhang Y, Yang J. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in gastric carcinoma using bioinformatics analysis. Med Sci Monit. 2019;25:8777–8796. doi:10.12659/MSM.916902
13. Wang B, Yin H, Zhang H, Wang T. circNRIP1 facilitates keloid progression via FXR1-mediated upregulation of miR‑503‑3p and miR‑503‑5p. Int J Mol Med. 2021;47:5. doi:10.3892/ijmm.2021.4903
14. Liu F, Li T, Zhan X. Silencing circular RNAPTPN12 promoted the growth of keloid fibroblasts by activating Wnt signaling pathway via targeting microRNA-21-5p. Bioengineered. 2022;13(2):3503–3515. doi:10.1080/21655979.2022.2029108
15. Zhang L, Tao H, Li J, et al. Comprehensive analysis of the competing endogenous circRNA-lncRNA-miRNA-mRNA network and identification of a novel potential biomarker for hepatocellular carcinoma. Aging. 2021;13(12):15990–16008. doi:10.18632/aging.203056
16. Lv W, Liu S, Zhang Q, et al. Circular RNA CircCOL5A1 sponges the MiR-7-5p/Epac1 axis to promote the progression of keloids through regulating PI3K/Akt signaling pathway. Front Cell Dev Biol. 2021;9:626027. doi:10.3389/fcell.2021.626027
17. Shi J, Yao S, Chen P, et al. The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol Biol Rep. 2020;47(1):201–209. doi:10.1007/s11033-019-05120-y
18. Wu F, He H, Chen Y, et al. CircPDE7B/miR-661 axis accelerates the progression of human keloid fibroblasts by upregulating fibroblast growth factor 2 (FGF2). Mol Cell Biochem. 2022;477(4):1113–1126. doi:10.1007/s11010-021-04345-5
19. Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi:10.1016/j.biomaterials.2013.11.083
20. Munagala R, Aqil F, Jeyabalan J, et al. Exosome-mediated delivery of RNA and DNA for gene therapy. Cancer Lett. 2021;505:58–72. doi:10.1016/j.canlet.2021.02.011
21. Hu JL, Wang W, Lan XL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. doi:10.1186/s12943-019-1019-x
22. Gupta R, Radicioni G, Abdelwahab S, et al. Intercellular communication between airway epithelial cells is mediated by exosome-like vesicles. Am J Respir Cell Mol Biol. 2019;60(2):209–220. doi:10.1165/rcmb.2018-0156OC
23. Li Q, Fang L, Chen J, et al. Exosomal MicroRNA-21 promotes keloid fibroblast proliferation and collagen production by inhibiting Smad7. J Burn Care Res. 2021;42(6):1266–1274. doi:10.1093/jbcr/irab116
24. Yuan R, Dai X, Li Y, Li C, Liu L. Exosomes from miR-29a-modified adipose-derived mesenchymal stem cells reduce excessive scar formation by inhibiting TGF-β2/Smad3 signaling. Mol Med Rep. 2021;24(5). doi:10.3892/mmr.2021.12398
25. Sokolova V, Ludwig AK, Hornung S, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi:10.1016/j.colsurfb.2011.05.013
26. Santoro D, Archer L, Chong E. Evaluation of cutaneous and circulating (serum and exosomes) levels of chemokines (CCL17, CCL22, CCL27 and CCL28) in atopic dogs and their correlation with severity of the disease. Vet Dermatol. 2022;33(3):195–e156. doi:10.1111/vde.13061
27. Abdelsaid K, Sudhahar V, Harris RA, et al. Exercise improves angiogenic function of circulating exosomes in type 2 diabetes: role of exosomal SOD3. FASEB J. 2022;36(3):e22177. doi:10.1096/fj.202101323R
28. Liu J, Peng X, Liu Y, et al. The diagnostic value of serum exosomal has_circ_0000615 for breast cancer patients. Int J Gen Med. 2021;14:4545–4554. doi:10.2147/IJGM.S319801
29. Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–D112. doi:10.1093/nar/gkx891
30. Wang J, Wu H, Xiao Z, Dong X. Expression profiles of lncRNAs and circRNAs in keloid. Plast Reconstr Surg Glob Open. 2019;7(6):e2265. doi:10.1097/GOX.0000000000002265
31. Jiao H, Ji G, Luo B, Chen C. CircCOL5A1 inhibits proliferation, migration, invasion, and extracellular matrix production of keloid fibroblasts by regulating the miR-877-5p/EGR1 axis. Burns. 2022. doi:10.1016/j.burns.2021.12.013
32. Gao H, Hu Z, Zhang X. Circular RNA hsa_circ_0057452 facilitates keloid progression by targeting the microRNA-1225-3p/AF4/FMR2 family member 4 axis. Bioengineered. 2022;13(5):13815–13828. doi:10.1080/21655979.2022.2084460
33. Marty P, Chatelain B, Lihoreau T, et al. Halofuginone regulates keloid fibroblast fibrotic response to TGF-β induction. Biomed Pharmacother. 2021;135:111182. doi:10.1016/j.biopha.2020.111182
34. Xie F, Teng L, Xu J, et al. Adipose-derived mesenchymal stem cells inhibit cell proliferation and migration and suppress extracellular matrix synthesis in hypertrophic-scar and keloid fibroblasts. Exp Ther Med. 2021;21(2):139. doi:10.3892/etm.2020.9571
35. Zhou R, Liao J, Cai D, et al. Nupr1 mediates renal fibrosis via activating fibroblast and promoting epithelial-mesenchymal transition. FASEB J. 2021;35(3):e21381. doi:10.1096/fj.202000926RR
36. Cui J, Jin S, Jin C, Jin Z. Syndecan-1 regulates extracellular matrix expression in keloid fibroblasts via TGF-β1/Smad and MAPK signaling pathways. Life Sci. 2020;254:117326. doi:10.1016/j.lfs.2020.117326
37. Thielitz A, Vetter RW, Schultze B, et al. Inhibitors of dipeptidyl peptidase IV-like activity mediate antifibrotic effects in normal and keloid-derived skin fibroblasts. J Invest Dermatol. 2008;128(4):855–866. doi:10.1038/sj.jid.5701104
38. Lei R, Li J, Liu F, et al. HIF-1α promotes the keloid development through the activation of TGF-β/Smad and TLR4/MyD88/NF-κB pathways. Cell Cycle. 2019;18(23):3239–3250. doi:10.1080/15384101.2019.1670508
39. Chen H, Xu X, Lai L, Huo R, Chen M. Circ_0008450 downregulates Runx3 to promote the proliferation and epithelial-mesenchymal transition of human keratinized epithelial cells. Cell Cycle. 2020;19(23):3303–3316. doi:10.1080/15384101.2020.1842665
40. Cui Y, Zheng Y, Lu Y, et al. LINC01224 facilitates the proliferation and inhibits the radiosensitivity of melanoma cells through the miR-193a-5p/ NR1D2 axis. Kaohsiung J Med Sci. 2022;38(3):196–206. doi:10.1002/kjm2.12467
41. Caramuta S, Egyházi S, Rodolfo M, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130(8):2062–2070. doi:10.1038/jid.2010.63
42. Polini B, Carpi S, Doccini S, et al. Tumor suppressor role of hsa-miR-193a-3p and −5p in cutaneous melanoma. Int J Mol Sci. 2020;21(17):6183. doi:10.3390/ijms21176183
43. Fang C, Dai CY, Mei Z, et al. microRNA-193a stimulates pancreatic cancer cell repopulation and metastasis through modulating TGF-β2/TGF-βRIII signalings. J Exp Clin Cancer Res. 2018;37(1):25. doi:10.1186/s13046-018-0697-3
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.