Back to Journals » Veterinary Medicine: Research and Reports » Volume 13
Isolation and Molecular Detection of Pasteurellosis from Pneumonic Sheep in Selected Areas of Amhara Region, Ethiopia: An Implication for Designing Effective Ovine Pasteurellosis Vaccine
Authors Akane AE, Alemu G , Tesfaye K, Ali DA , Abayneh T, Kenubih A , Ejo M , Shite Abat A , Admassu B, Ibrahim SM
Received 6 March 2022
Accepted for publication 14 April 2022
Published 24 April 2022 Volume 2022:13 Pages 75—83
DOI https://doi.org/10.2147/VMRR.S365267
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Young Lyoo
Aragaw Ebabu Akane,1 Gashaw Alemu,2 Kidest Tesfaye,2 Destaw Asfaw Ali,2 Takele Abayneh,3 Ambaye Kenubih,2 Mebrat Ejo,4 Anmaw Shite Abat,2 Bemrew Admassu,4 Saddam Mohammed Ibrahim2
1Lalibela Town Administration Agricultural Office, Lalibela, Ethiopia; 2Department of Veterinary Paraclinical Studies, College of Veterinary Medicine and Animal Sciences, University of Gondar, Gondar, Ethiopia; 3National Veterinary Institute, Bishoftu, Ethiopia; 4Department of Veterinary Biomedical Sciences, College of Veterinary Medicine and Animal Sciences, University of Gondar, Gondar, Ethiopia
Correspondence: Saddam Mohammed Ibrahim, Email [email protected]
Introduction: Pneumonic pasteurellosis mainly caused by bacterial species of Mannheimia, Pasteurella, and Bibersteinia causes a significant financial loss to the sheep production sector through reduced productivity and high mortality. There is a dearth of information on the major agents involved in the disease in the Amhara region, Ethiopia. Therefore, the aim of this study was to isolate and molecularly confirm Mannheimia, Pasteurella, and Bibersteinia from nasal swabs of sheep suspected of pneumonic pasteurellosis in selected areas of the Amhara region.
Methods: Isolation and phenotypic characterization were performed using microbiological and biochemical testing according to standard methods. Molecular confirmation of isolates was done through amplification of virulence associated genes, PHSAA and Rpt2, of Mannheimia hemolyticausing multiplex PCR.
Results: Accordingly, 46 out of 141 (32.62%) samples were presumably identified as M. hemolytica with no Pasteurella multocida and Bibersteinia trehalosi. Seven (n=7) out of the 46 isolates tested positive for either of the two virulence genes.
Discussion and conclusion: The finding of this study is indicative that M. hemolytica is the main bacteria linked with pneumonic pasteurellosis in the study area which suggests the need to develop a polyvalent vaccine including strains of M. hemolytica or its antigenic determinants. However, the role of other bacterial, viral, and parasitic agents in the cases investigated should also be considered.
Keywords: Bibersteinia trehalosi, isolation, Mannheimia hemolytica, Pasteurella multocida, PHSSA, pneumonic pasteurellosis, Rpt2, sheep
Introduction
Ethiopia is known for its high and diverse sheep population with an estimate of 43 million sheep, of which 10.4 million are found in the Amhara region, one of the administrative regions in the country.1 In addition to serving as source of immediate cash, sheep production provides an insurance in instances of low crop productivity due to erratic rainfall, severe erosion, frost, and water logging problems.2,3 Thus, increasing the current level of sheep productivity is essential to meet the demands of the ever-increasing human population in the country.4
Of the several threats to the production sector, pneumonic pasteurellosis is a major concern for being economically devastating by causing reduced live weight, delayed marketing, increased treatment cost, and loss of animals in cases of outbreaks.5 Clinically, an acute infectious disease is triggered when the immune system of animals is compromised by stress factors such as overcrowding, transportation, drought, extreme weather and other concurrent disease.6 The main causative agents of the disease are Pasteurella multocida, Mannheimia hemolytica, and Bibersteinia trehalosi, commensals of the upper respiratory tract of sheep, which descend to the lower respiratory tract when animals become immunosuppressed.7–9
Vaccination with inactivated P. multocida biotype A vaccine (National Veterinary Institute, Ethiopia) is currently practiced nationally on an annual basis to curb the impact of the disease. Nevertheless, outbreaks of pneumonic pasteurellosis are reported yearly in different parts of the country including the Amhara region. Among the possible reasons for limited vaccine efficacy, is a mismatch between the vaccinal and bacterial strains involved, particularly, when immunity against the disease is serotype-specific.10–12 Despite few reports based on prospective clinical or bacteriological examinations, there are no studies conducted to isolate and molecularly confirm the causative agents involved in such types of outbreaks and supposed cases of pneumonic pasteurellosis in the Amhara region. Determination and characterization of the major pathogens involved is an essential input for vaccine producers to formulate effective vaccines.13 Therefore, the aim of this study was to isolate and molecularly detect M. hemolytica, P. multocida, and B. trehalosi from pneumonic sheep.
Materials and Methods
Study Area
The study was conducted in selected districts and towns of central, north, and south Gondar zones of the Amhara region, which includes Gondar town, Debark town, Dembia, Gondar Zuria, Wegera, and Farta districts (Figure 1).
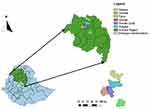 |
Figure 1 Map of the study sites. Amhara region, northwest of Ethiopia and the various towns and districts within the Amhara region included in the study. |
Study Animals
Animals included in this study were sheep presented to the veterinary clinics found in the study sites having signs of respiratory problems suspected as pneumonic pasteurellosis. Therefore, sheep manifesting the clinical signs: coughing, dyspnea, lethargy, oculonasal discharges (serous to mucopurulent), and fever were considered for bacteriological sampling irrespective of their age, sex, breed, physiological, and vaccination status. Animals were handled and approached humanely, and have received the standard of veterinary care during their visits to the clinics.
Study Design and Sampling Technique
A cross-sectional study design and purposive method of sampling was employed with the objective of isolating and identifying M. hemolytica, P. multocida, B. trehalosi from sheep suspected of having pneumonic pasteurellosis from November 2019 to May 2020. The study districts were selected purposively based on prior information on the problem and availability of sheep population. A total of 141 sheep fulfilling the clinical diagnostic criteria were sampled from the different study sites as follows: Gondar town (n=24), Debark town (n=25), Dembia district (n=27), Gondar zuriya district (n=10), Wegera district (n=23), and Farta district (n=32).
Sampling Procedure
During sample collection each animal was individually identified and restrained by an assistant. External nares were decontaminated using a cotton soaked with 70% ethanol prior to sampling.
Next, sterile cotton-tipped swabs moistened with tryptone soy broth (Oxoid, Hampshire, England) were inserted into the nasal cavities of each sheep and the mucus surface was rubbed by rotating the swabs on both nostrils gently. The swabs were allowed to remain in contact with the secretions for up to one minute and were placed back into labeled sterile universal tubes containing 3 mL of sterile tryptone soy broth (Oxoid). Labeled samples of nasal swabs were transported in icebox to the veterinary microbiology laboratory of the University of Gondar (UoG), College of Veterinary Medicine and Animal Science (CVMAS) for microbiological analysis. Samples were either processed immediately upon arrival or maintained and preserved at +4°C until further process.14
Bacteriological Assessment
Nasal swabs were incubated in tryptone soy broth (Oxoid) for 24 h at 37°C after which they were streaked into blood agar containing 5% defibrinated sheep blood (Titan Biotech Limited, Bhiwadi, India). Blood agar plates were then incubated at 37°C for 48 h. Subsequently, from culture-positive plates, typical colonies were Gram stained to determine staining property and cellular morphology under light microscope. Those colonies showing a Gram reaction and colony characteristics indicative of Pasteurella were subcultured on blood and MacConkey agar for further analysis. The presence of growth in blood agar without hemolysis and lack of growth in MacConkey is considered suggestive of P. multocida. Colonies were presumptively identified to be M. hemolytica and B. trehalosi, when they grow on blood agar (round, translucent and grayish colonies) with typical ß-hemolytic pattern, and grow on MacConkey agar appearing pink pinpointed colonies, an indicator of lactose fermentation.15
Biochemical Identification
Pure cultures of single colony type from both blood and MacConkey agars were subcultured on nutrient agar-slants at 37°C for 24 h for a series of primary and secondary biochemical tests. The biochemical tests were done as described previously.15 Finally, colonies presumably identified as M. hemolytica were subcultured on 8 mL nutrient broth and preserved using 98% 2 mL glycerol for transportation into NVI, Bishoftu, Ethiopia for molecular identification.
Molecular Detection
Extraction of Bacterial Deoxyribose Nucleic Acid
Bacterial DNA extraction was conducted at the molecular biology laboratory of NVI, Ethiopia. A few colonies from the phenotypically characterized pure cultures of M. hemolytica grown on nutrient agar for 24 to 48 h were transferred into 1.8 mL Eppendorf tubes.16 Bacterial DNA was extracted using Qiagen DNeasy Blood and Tissue Kit as per manufacturer’s instructions (Qiagen, German town, MD, USA).
Detection of Virulence Genes of M. hemolytica Using Multiplex PCR
Multiplex PCR (mPCR) was performed by simultaneously amplifying the two virulence-associated genes of M. hemolytica, i.e .PHSSA (P. hemolytica serotype specific antigen), a serotype specific antigen of M. hemolytica, and Rpt2, a gene coding for methyltransferase as reported previously.16 The primer design and size of amplified products is provided in Table 1. The reaction mixture for the PCR comprised: 10 μL of IQ Supermix (Bio Rad, USA) (DNA polymerase, dNTPs and buffer), 2 μL (5 pM/μL) of each primer pairs, 3 μL of nuclease-free water and 4 μL DNA template, totaling a reaction volume of 25 μL. Regarding the PCR condition, initial denaturation was performed at 95°C for 3 min, after which 35 cycles of each at 95°C for 1 min were completed. Then, annealing took place at 48°C for 1 min followed by an initial extension at 72°C for 30 seconds, and a final extension cycle at 72°C for 5 min.16 Three reaction tubes: with DNA extraction control, without the DNA template (nuclease-free water), and with a DNA template from reference M. hemolytica from NVI culture pool (MH-NVI) were included as extraction, negative and positive controls respectively.
 |
Table 1 Primers Sequences Used in the Amplification of PHSSA and Rpt2 in Molecular Detection of M. hemolytica |
Visualization of PCR Products Through Agarose Gel Electrophoresis
Agarose gel (2% w/v, 0.5× Tris borate EDTA buffer) stained with gel red was utilized in the analysis and detection of the PCR products. Each PCR product (5 µL) were mixed with 6× loading dye and loaded into a separate well of the preprepared gel while 1 kb plus DNA molecular markers were loaded onto the first and last lane and run at 120 V for 60 min on electrophoresis apparatus (EC 2060, USA). Finally, the different bands were visualized under UV transilluminator and photographed in a gel documentation system (UVI TEC, UK).
Data Management and Analysis
Data collected were coded and entered into the Microsoft Excel spreadsheet. Frequency and percentages were used to describe the isolation rate and the distribution across different study sites.
Results
Bacteriological and Biochemical Characteristics of Isolates
In this study, M. hemolytica was found to be the predominantly identified bacterial species (32.62%, 46/141) from cases of ovine pneumonic pasteurellosis throughout the study areas. Isolation rates as per study sites were as followed: 20.83% (5/24), 29.6% (8/27), 2% (2/10), 39.1% (9/23), 36% (9/25), and 40.6% (13/32) for Gondar town, Debark town, Dembia, Gondar Zuria, Wegera, and Farta districts, respectively (Table 2).
 |
Table 2 Frequency and Isolation Rate of Presumptive M. hemolytica in Different Study Areas |
Isolates phenotypically identified as M. hemolytica were gram-negative, short ovoid rods arranged in chains with an occasional tendency of bipolar staining. In addition, these isolates were able to grow on blood agar with complete zones of hemolysis. Growth on MacConkey agar was also observed with varying degree of lactose fermentation evident by pinpoint red colonies. Biochemically, isolates were positive for catalase and oxidase while testing negative for methyl red, urease, citrate, and indole tests. Sugar fermentation profile indicated that the isolates were able to metabolize different sugars such as arabinose, maltose, sucrose, glucose, sorbitol and lactose but not trehalose. In this study, P. multocida and B. trehalosi were not identified as none of the colonies fulfilled the characteristic growth of these species on MacConkey and blood agar, and on the variety of biochemical tests as well (Table 3).
 |
Table 3 Biochemical Characteristics of Isolates from Cultures of Nasal Swabs of Sheep Affected with Pneumonic Pasteurellosis |
Molecular Confirmation of Isolates
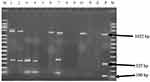
All of the isolates (n=46) presumably identified as M. hemolytica through microbiological and biochemical analysis were further subjected to mPCR test, amplifying PHSSA and Rpt2 genes. Seven (n=7) out of the 46 isolates (15.21%) tested positive for either of the genes, i.e. three (n=3) of them were positive for both PHSSA (serotype A1) and Rpt2 genes; two (n=2) of them tested positive only for Rpt2 gene; while the remaining samples (n=2) tested positive only for PHSSA gene (Figure 2).
Discussion
Ovine pasteurellosis causes a significant harm to the sheep production sector affecting the livelihood of communities that rely on sheep rearing as well as industries that process sheep meat and their products at large.17 Pasteurella multocida and Mannheimia hemolytica are the main causative agents of the disease responsible for 30% of deaths in feedlot cattle and acute outbreaks in sheep population with consequence of massive mortality all across the globe.9 Several biotypes and serotypes of these bacterial species have been reported with varied distribution across different areas.6,18 The aim of this study was to isolate Pasteurella, Mannheimia, and Bibersteinia involved in pneumonic pasteurellosis of sheep in selected areas of the northwest Amhara region, Ethiopia. The finding of this study indicated that M. hemolytica is the major bacteria associated with cases of pneumonic pasteurellosis in the study areas. Bacteriological and biochemical methods were employed in the isolation and presumptive identification of the bacteria species. Of the 141 cultures of nasal swabs, 46 (32.62%) were tentatively identified as M. hemolytica. The microbiological and biochemical features of the isolates, and the clinical feature of affected sheep in the present study are consistent with M. hemolytica and pneumonic pasteurellosis described elsewhere.19,20
This finding agrees with previous studies, which reported that M. hemolytica is the most common agent isolated in cases of ovine pneumonic pasteurellosis.13,21,22 The recovery rate of M. hemolytica in this study is lower than reports of Legesse et al (2018)13 in central Ethiopia (34.21%): Aschalew (1998)23 in North Shewa (35%): Mekonnen24 in Arsi (48%): Alemneh et al25 in Fogera wereda (79.5%): Abera et al26 in Bedelle districts (46%): and Kaoud et al27 in Egypt (52%). This variation could arise from various factors including difference in sample size and season of sample collection, geographical variation, site of sampling, and sensitivity of tests used. In fact, Ali and Al Balaa28 reported a significant increase in isolation rate of M. hemolytica in summer and winter as compared to spring. Contrary to our findings, Marru et al,21 Deressa et al,22 Abera et al,26 and Tesfaye29 reported to isolate either B. trehalose or P. multocida or both, none of which were recovered in the present study. Similarly, variation in sample size, geographical areas, animal factors (age for example), and management practice between the study sites could explain why those species were not isolated in the current study. Of note, Alemneh et al25 reported a significantly higher frequency of infection with pneumonic pasteurellosis in winter and spring compared to autumn and summer. Age-wise, B. trehalosi is reported to cause a septicemic disease in young lambs which often emerges in the form of severe outbreaks.30 The application of monovalent vaccine derived from P. multocida biotype A (NVI, Ethiopia) in our study areas may have lessened the contribution of P. multocida, thus, its isolation from diseased animals.
In this study molecular confirmation of M. hemolytica was performed though mPCR, simultaneously amplifying the genes, PHSSA and Rpt2. Seven (n=7) out of the 46 the isolates were confirmed to harbor those genes, which is contrary to the studies of Legesse et al13 and Hawari et al31 which reported that all isolates identified to be M. hemolytica through microbiological and biochemical tests were all found to be positive for PCR detection.
The serotype specific antigen, PHSSA, is reported to play a role in conversion of commensal microbes into pathogenic in stressful situations indicating its involvement in pathogenesis of pneumonic pasteurellosis due to M. hemolytica.16,32 In contrast, the species-specific Rpt2 gene, coding for methyltransferase, plays an important role in protection of bacterial DNA from endonuclease cleavage by catalyzing the methylation of a specific DNA recognition sequence designating it a self-DNA. Thus, Rpt2 takes part in modulation of type III restriction-modification system.16,33 Therefore, Rpt2 and PHSSA genes of M. hemolytica are appropriate molecular diagnostic targets.16 As mentioned earlier, isolates (n=39) passing the phenotypic criteria of M. hemolytica tested negative for both of the virulence-associated genes. These isolates are likely to belong to serotypes other than A1 (as the PHSSA used was of A1 serotype) or nonpathogenic strains of M. hemolytica.34
Ovine pasteurellosis is a disease of multifactorial etiological agents. In addition to the external factors, the presence of different bacterial biotypes and serotypes with immunity that does not cross protect further complicates the problem. As mentioned earlier, presently, a monovalent vaccine based on P. hemolytica biotype A (NVI, Bishoftu, Ethiopia) is used in Ethiopia. Infection with one bacterial species or serotype does not confer protection against infection or disease by other bacterial species or serotypes.10–12 Thus, vaccination with a monovalent P. hemolytica biotype A based vaccine is unlikely to provide adequate protection against diseases caused by M. hemolytica which is indicated to be the prevailing agent associated with pneumonic pasteurellosis in the study areas. Therefore, this suggests the need to develop a multivalent vaccine that incorporates appropriate bacterial strains of M. hemolytica or it’s antigenic determinants for enhanced vaccine effectiveness and disease control strategies.
Conclusion
The current study reports the first attempt to isolate and molecularly detect M. hemolytica from pneumonic sheep in selected areas of the Amhara region, Ethiopia. Our finding is indicative of the significant association of M. hemolytica with pneumonic pasteurellosis in the study area. The current vaccination strategy using inactivated P. hemolytica biotype A vaccine could provide little-to-no protection against outbreaks of pneumonic pasteurellosis that may involve diverse serotypes of M. hemolytica, which may call for the need to develop a polyvalent vaccine including M. hemolytica or its antigenic determinants. This, in turn, could augment other control and prevention strategies in the region, thus, avoiding the drastic consequences of the disease. However, the study did not cover significant other parts of the region, which entails further studies to identify and molecularly (phylogenetically) characterize the etiological agents implicated in the disease. Finally, the involvement of other potential pathogens in the respiratory cases investigated in this study needs to be taken into consideration.
Abbreviations
CVMAS, College of Veterinary Medicine and Animal Sciences; DNA, deoxyribonucleic acid; mPCR, multiplex polymerase chain reaction; NVI, National Veterinary Institute; PHSSA, P. hemolytica serotype specific antigen; UoG, University of Gondar.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon a formal request.
Ethical Approval and Consent to Participation
This study has obtained an ethical approval by institutional Review Board of UoG, CVMAS. Animals were approached with great care according to the guidelines for ethics of animal research. Moreover, animal owners were consented, and the benefits and outcomes of the study explained for the study participants.
Acknowledgments
We are grateful to laboratory technicians at UoG, CVMAS, animal owners, and district veterinarians for their invaluable contribution to the success of this study. The mPCR was conducted in NVI, Bishoftu, Ethiopia, with the generous support of the institute which we would like to highly appreciate.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work is part of a mega research project funded by the University of Gondar, office of the vice president for research and community service (Budget code: VPRCS 6223, 2019/20-2020/21) to SMI as PI and the co-authors as co-investigators of the project.
Disclosure
The authors report no conflicts of interest in this work.
References
1. CSA. Agricultural Sample Survey. Addis Ababa: Ethiopia: Central Statistical Authority; 2020.
2. Zygoyiannis D. Sheep production in the world and in Greece. Small Rumin Res. 2006;62(1–2):143–147. doi:10.1016/j.smallrumres.2005.07.043
3. Tibbo M, Woldemeskel M, Gopilo A. An outbreak of respiratory disease complex in sheep in central Ethiopia. Trop Anim Health Prod. 2001;33(5):355–365. doi:10.1023/a:1010565905004
4. Getachew T, Haile A, Tibbo M, Sharma AK, Sölkner J, Wurzinger M. Herd management and breeding practices of sheep owners in a mixed crop-livestock and a pastoral system of Ethiopia. Afr J Agric Res. 2010;5(8):685–691. doi:10.5897/AJAR10.392
5. Zheng T, Gupta SK, McCarthy AR, Moffat J, Buddle BM. Cross-protection study of a Mannheimia haemolytica serotype 1 vaccine against acute pasteurellosis in lambs induced by a serotype 2 strain. Vet Microbiol. 2015;177(3–4):386–393. doi:10.1016/j.vetmic.2015.02.019
6. Mohamed RA, Abdelsalam EB. A review on pneumonic pasteurellosis (respiratory mannheimiosis) with emphasis on pathogenesis, virulence mechanisms and predisposing factors. Bulg J Vet Med. 2008;11(3):139–160.
7. Hounsome JD, Baillie S, Noofeli M, et al. Outer membrane protein A of bovine and ovine isolates of Mannheimia haemolytica is surface exposed and contains host species-specific epitopes. Infect Immun. 2011;79(11):4332–4341. doi:10.1128/IAI.05469-11
8. García-Alvarez A, Fernandez-Garayzabal JF, Chaves F, Pinto C, Cid D. Ovine Mannheimia haemolytica isolates from lungs with and without pneumonic lesions belong to similar genotypes. Vet Microbiol. 2018;219:80–86. doi:10.1016/j.vetmic.2018.04.012
9. Sahay S, Natesan K, Prajapati A, et al. Prevalence and antibiotic susceptibility of Mannheimia haemolytica and Pasteurella multocida isolated from ovine respiratory infection: a study from Karnataka, Southern India. Vet World. 2020;13(9):1947. doi:10.14202/vetworld.2020.1947-1954
10. Fodor L, Pénzes Z, Varga J. Coagglutination test for serotyping Pasteurella haemolytica. J Clin Microbiol. 1996;34(2):393–397. doi:10.1128/jcm.34.2.393-397.1996
11. Purdy CW, Cooley JD, Straus DC. Cross-protection studies with three serotypes of Pasteurella haemolytica in the goat model. Curr Microbiol. 1998;36(4):207–211. doi:10.1007/s002849900295
12. Odugbo MO, Odama LE, Umoh JU, Lombin LH. The comparative pathogenicity of strains of eight serovars and untypable strains of Mannheimia haemolytica in experimental pneumonia of sheep. Vet Res. 2004;35(6):661–669. doi:10.1051/vetres:2004044
13. Legesse A, Abayneh T, Mamo G, et al. Molecular characterization of Mannheimia haemolytica isolates associated with pneumonic cases of sheep in selected areas of Central Ethiopia. BMC Microbiol. 2018;18(1):1–10. doi:10.1186/s12866-018-1338-x
14. Carter GR, Cole JR
15. Quinn PJ, Morkey BK, Carter ME, Donnelly WJC, Lenard FC, Maguire D. Pasteurella species and Mannheimia haemolytica. In: Veterinary Microbiology and Microbial Diseases.
16. Kumar J, Dixit SK, Kumar R. Rapid detection of Mannheimia haemolytica in lung tissues of sheep and from bacterial culture. Vet World. 2015;8(9):1073. doi:10.14202/vetworld.2015.1073-1077
17. Hussain R, Mahmood F, Ali HF, Siddique AB. Bacterial, PCR and clinico-pathological diagnosis of naturally occurring pneumonic pasteurellosis (mannheimiosis) during subtropical climate in sheep. Microb Pathog. 2017;112:176–181. doi:10.1016/j.micpath.2017.09.061
18. Ilhan Z, Keleş I. Biotyping and serotyping of Mannheimia (Pasteurella) haemolytica isolated from lung samples of slaughtered sheep in the Van region. Turk J Vet Anim Sci. 2007;31(2):137–141.
19. Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, Fitz Patrick ES. Veterinary Microbiology and Microbial Disease.
20. Gilmour N. Pasteurella haemolytica infections in sheep. Vet Q. 1980;2(4):191–198. doi:10.1080/01652176.1980.9693780
21. Marru HD, Anijajo TT, Hassen AA. A study on Ovine pneumonic pasteurellosis: isolation and identification of Pasteurellae and their antibiogram susceptibility pattern in Haramaya District, Eastern Hararghe, Ethiopia. BMC Vet Res. 2013;9(1):1–8. doi:10.1186/1746-6148-9-239
22. Deressa A, Asfaw Y, Lubke B, Kyule MY, Tefera G, Zessin KH. Molecular detection of Pasteurella multocida and Mannheimia haemolytica in sheep respiratory infections in Ethiopia. Intern J Appl Res Vet Med. 2010;8(2):101.
23. Aschalew Z. A study on pneumonic pasteurellosis in North Shewa [DVM thesis]. Debre-Zeit: Ethiopia: FVM, Addis Ababa University; 1998:1–59.
24. Mekonnen T. An epidemiological study on ovine pasteurellosis in Arsi, Southeast Ethiopia [DVM Thesis]. Debre-Zeit: Addis Ababa University, Faculty of Veterinary Medicine; 2000.
25. Alemneh T, Tewodros A. Sheep and goats pasteurellosis: isolation, identification, biochemical characterization and prevalence determination in Fogera Woreda, Ethiopia. J Cell Anim Biol. 2016;10(4):22–29. doi:10.5897/JCAB2016.0449
26. Abera D, Sisay T, Birhanu T. Isolation and identification of Mannheimia and Pasteurella species from pneumonic and apparently healthy cattle and their antibiogram susceptibility pattern in Bedelle District, Western Ethiopia. J Bacteriol Res. 2014;6(5):32–41. doi:10.5879/JBR2014.0143
27. Kaoud H, El-Dahshan AR, Zaki MM, Nasir SA. Occurrence of Mannheimia haemolytica and Pasteurella trehalosi among ruminants in Egypt. NY Sci J. 2010;3(5):135–141.
28. Ali H, Al Balaa B. Prevalence of Mannheimia haemolytica in Syrian Awassi Sheep. Bulg J Vet Med. 2019;22(4):439–446. doi:10.15547/bjvm.2123
29. Tesfaye S. Serological and bacteriological investigation of P. haemolytica serotypes in sheep in the high lands of wollo, North Eastern Ethiopia [DVM Thesis]. Debre-Zeit, Ethiopia: AAU, FVM; 1997:1–65.
30. Szeredi L, Rausch F, Szeleczky Z, Janosi S. High mortality caused by Bibersteinia trehalosi septicaemia in adult sheep–A case report. Acta Vet Hung. 2018;66(4):509–517. doi:10.1556/004.2018.045
31. Hawari AD, Hassawi DS, Sweiss M. Isolation and identification of Mannheimia haemolytica and Pasteurella multocida in sheep and goats using biochemical tests and random amplified polymorphic DNA (RAPD) analysis. J Biol Sci. 2008;8(7):1251–1254. doi:10.3923/jbs.2008.1251.1254
32. Rawat N, Gilhare VR, Kushwaha KK, et al. Isolation and molecular characterization of Mannheimia haemolytica and Pasteurella multocida associated with pneumonia of goats in Chhattisgarh. Vet World. 2019;12(2):331. doi:10.14202/vetworld.2019.331-336
33. Ryan KA, Lo RY. Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res. 1999;27(6):1505–1511. doi:10.1093/nar/27.6.1505
34. Rice JA, Medina-Carrasco L, Hodgins DC, et al. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev. 2007;8(2):117–128. doi:10.1017/S1466252307001375
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.