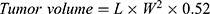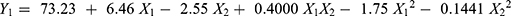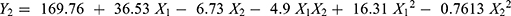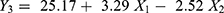Back to Journals » International Journal of Nanomedicine » Volume 17
Investigating the Impact of Optimized Trans-Cinnamic Acid-Loaded PLGA Nanoparticles on Epithelial to Mesenchymal Transition in Breast Cancer
Authors Badawi NM, Attia YM, El-Kersh DM, Hammam OA , Khalifa MKA
Received 23 October 2021
Accepted for publication 13 January 2022
Published 18 February 2022 Volume 2022:17 Pages 733—750
DOI https://doi.org/10.2147/IJN.S345870
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lijie Grace Zhang
Noha M Badawi,1 Yasmeen M Attia,2 Dina M El-Kersh,3 Olfat A Hammam,4 Maha KA Khalifa5
1Department of Pharmaceutics and Pharmaceutical Technology, Faculty of Pharmacy, The British University in Egypt, Cairo, Egypt; 2Department of Pharmacology, Faculty of Pharmacy, The British University in Egypt, Cairo, Egypt; 3Department of Pharmacognosy, Faculty of Pharmacy, The British University in Egypt, Cairo, Egypt; 4Department of Pathology, Theodor Bilharz Research Institute, Cairo, Egypt; 5Department of Pharmaceutics and Pharmaceutical Technology, Faculty of Pharmacy (Girls), Al-Azhar University, Cairo, Egypt
Correspondence: Noha M Badawi, Department of Pharmaceutics and Pharmaceutical Technology, Faculty of Pharmacy, The British University in Egypt, Suez Desert Road, P.O. Box 11562, El Sherouk City, Cairo, Egypt, Email [email protected]
Purpose: To design and optimize trans-cinnamic acid-loaded PLGA nanoparticles (CIN-PLGA-NPs) and assess its inhibitory effect on epithelial-mesenchymal transition (EMT) in triple-negative breast cancer.
Methods: The quality by design approach was used to correlate the formulation parameters (PLGA amount and Poloxamer188 concentration) and critical quality attributes (entrapment efficiency percent, particle size and zeta potential). Design of CIN-PLGA-NPs formulations was done based on central composite response surface design and formulated by nanoprecipitation method. In addition, the optimized CIN-PLGA-NPs formulation was further evaluated for morphology using transmission electron microscopy and in vitro dissolution test. The cytotoxicity of CIN-PLGA-NPs optimized formula in comparison to the free trans-cinnamic acid (CIN-Free) was investigated in vitro using MDA-MB-231, triple-negative breast cancer cells, followed by scratch wound assay for evaluating the impact on the migratory potential of MDA-MB-231 cells. In vivo antitumor activity was evaluated using Ehrlich ascites carcinoma solid tumor animal model where tumor volumes were measured at different time points and necrotic/apoptotic indices were estimated in tumor sections. EMT markers, E- and N-cadherin, were assessed in solid tumors as well.
Results: The optimized formulation showed entrapment efficiency of 76.98%, particle size of 186.3 nm with a smooth spherical surface and zeta potential of − 28.47 mV indicating its stability. Furthermore, CIN-PLGA-NPs optimized formula released 60.8± 1.89% of the total CIN-Free within 24 hours compared to 29± 1.25% of the raw CIN-Free indicating improved dissolution rate. The optimized formula showed superior cytotoxicity on MDA-MB-231 cells compared to its free counterpart as well as increased wound closure percentage along with reduced tumor size in mice and increased necrotic and apoptotic indices. Tumor levels of E-cadherin and N-cadherin were indicative of EMT inhibition.
Conclusion: Our findings proved the capability of PLGA nanoparticles in loading trans-cinnamic acid in addition to enhancing its antitumor efficacy in triple-negative breast cancer possibly via inhibiting EMT.
Keywords: trans-cinnamic acid, epithelial-mesenchymal transition, PLGA nanoparticles, quality by design, breast cancer
Introduction
Breast cancer is considered the most common reason of cancer mortality among women.1 It is classified into hormone receptor-positive, human epidermal growth factor receptor-2 (HER2) overexpressing and triple-negative breast cancer,2 the latter being clinically characterized by the negative expression of progesterone receptor (PR), estrogen receptor (ER), and HER2.3 Triple-negative breast cancer is considered the most aggressive and heterogeneous subtype of breast cancer stemming from its ability to grow rapidly and metastasize to the viscera, particularly the lungs and the brain. Therefore, its aggressive nature, metastatic potential, and lack of receptor/target expression presents conventional chemotherapy as the pillar for treating triple-negative breast cancer.3 Chemotherapy, however, is toxic to non-cancerous cells and frequently leads to rapid development of acquired resistance in tumor cells, owing to the upregulation of multidrug resistance (MDR) transporters.4 In addition, chemotherapy has a fast clearance, low therapeutic efficacy, uncontrolled drug release behavior, and lack of selectivity.5 Hence, there is an urgent need to develop alternative safer therapies to overcome a multitude of adverse effects affiliated to chemotherapy. Accordingly, the anticancer activities of natural products and their synthetic derivatives have been extensively studied, primarily as chemopreventive and chemotherapeutic agents.6
Epithelial to mesenchymal transition (EMT) is a dynamic process that is highly active in triple-negative breast cancer tumors where mesenchymal attributes become dominant over epithelial ones.7 This process adds to the aggressiveness of triple-negative breast cancer where the cells undergo phenotypic changes and start losing their polarity, cell-to-cell adhesion and differentiation increasing their stemness and, therefore, resistance to radio- and chemotherapy.8 Accordingly, targeting EMT has emerged as a promising treatment modality in cancer.9
Cinnamic acid is a natural aromatic carboxylic acid that is found in many plants for example Cinnamomum cassia (Chinese cinnamon) and Panax ginseng.10 Cinnamic acid exhibits either cis or trans configuration due to the presence of an acrylic acid group which can be substituted on the phenyl ring.10 The trans-cinnamic acid (CIN) is more stable than the cis-isomer which explains why it is the most naturally predominant form (>99%).11 Previous studies reported antimicrobial, antidiabetic, neuroprotective, anti-inflammatory, antioxidant and anticancer properties of CIN.10 Inhibition of DNA synthesis, suppression of nuclear factor kappa B (NF-kB) activation, generation of IL-8, inhibition of histone deacetylase and antioxidant activities are among the mechanisms behind CIN cytotoxic effects.12 However, the application of CIN is limited due to its poor stability, hydrophobicity and low oral bioavailability.13
Several studies, therefore, highlighted the fact that nanoparticle-based drug delivery systems might improve the solubility and stability of the encapsulated drugs while maintaining their intracellular therapeutic concentration.14 Furthermore, other characteristics such as increased drug loading ability, longer half-life with mostly minor systemic toxicity, increased internalization into the tumor via endocytosis, sustained and regulated release of cytotoxic drug over the suitable duration and time along with body excretion are significant for nanoparticles in cancer treatment.15 Biodegradable polymeric nanoparticles, lipid nanoparticles, magnetic nanoparticles, polymeric micelles, liposomes, niosomes, and nanoemulsions are all examples of nanotechnology formulation types.16 The major drawbacks of most of the nanoparticle-based systems include low shelf‐life stability, the use of high concentrations of surfactant and cosurfactant, the liability of lipid oxidation and transformation, incompatibility with various active agents, and a limited drug loading efficiency.17 In contrast, polymeric nanoparticles are more stable in vivo, with high drug loading capacities, and can release drugs in a controlled manner.18 Polymeric nanoparticles which also appeared to be the best-fit for drug delivery in cancer therapy owing to their higher affinity for favored accumulation in certain solid tumors through the leaky endothelial tissue surrounding the tumor by enhanced permeability and retention (EPR) effect. However, recently different approaches were adopted and successfully implemented to target either triple-negative breast cancer, in general, or EMT-induced chemoresistance using lysyl oxidase engineered lipid nanovesicles19 and emodin-loaded polymer-lipid hybrid nanoparticles,20 respectively.
Poly-lactide-co-glycolide (PLGA) is considered a good polymeric nanocarrier owing to its biodegradability, biocompatibility and extended drug release profiles.21 In addition, it has been adopted as a drug delivery system by the Food and Drug Administration (FDA) and the European Medicine Agency (EMA).14 PLGA nanoparticles are made up of hydrophobic groups on the inside and polar groups on the outside to enable effective encapsulation of both hydrophobic and hydrophilic drugs. Hydrophobic interactions and hydrogen bonding allow phenolic phytochemicals to interact with the hydrophobic sites of the nanoparticles. As a result of their inter-particle repulsions and hydration, nanoparticles (NPs) can remain stable in a dispersion system.22
A novel method, known as quality by design (QbD), has been approved by the FDA. It is a systematic approach for designing the final product based on certain quality necessities known as critical quality attributes (CQAs) of the formulation. The use of QbD ensures the quality of the final product by better understanding and implementing both process and formulation factors.23
To the best of our knowledge, there is no fabricated formulation of CIN-PLGA-NPs for the management of triple-negative breast cancer via inhibiting EMT. Therefore, our main aim was to formulate, characterize and optimize CIN-PLGA-NPs using QbD approach based on response surface central composite design where a correlation between formulation parameters (PLGA amount and Poloxamer188 concentration) and CQAs (entrapment efficiency percent, particle size and zeta potential) of the developed formulations were investigated. Furthermore, we studied the antitumor effect of the optimized formula in vitro using the triple-negative breast cancer cell line, MDA-MB-231, and in vivo using an animal model of Ehrlich ascites carcinoma (EAC) solid tumor in comparison to the CIN-Free. Further insights into the potential inhibitory effect of the optimized formula on EMT were also provided.
Materials and Methods
Materials
Trans-Cinnamic acid was purchased from Acros-Organics (USA). PLGA (50:50) was purchased from Sigma-Aldrich (USA). Poloxamer188 was kindly provided by the Egyptian International Pharmaceutical Industries Co., EIPICO (Egypt). Acetone (HPLC grade) was purchased from Fisher Scientific (UK). Dialysis tubing cellulose membrane (molecular weight cut off 12,000–14,000 Dalton) was purchased from Sigma-Aldrich (USA). All other materials were of analytical grade.
Methods
Preparation of CIN-PLGA-NPs
CIN-PLGA-NPs were fabricated using single emulsion solvent evaporation method (nanoprecipitation method). Through this method, the organic phase was prepared by dissolving precisely weighed PLGA and CIN in 10 mL of acetone as organic solvent. Afterwards, the organic phase was dropped into 20 mL of aqueous phase containing surfactant (Poloxamer188) at the rate of 1 mL/min. The nanoparticle suspension was subjected to continuous stirring at 1000 rpm overnight and kept at room temperature to allow the complete evaporation of acetone, resulting in the colloidal suspension of PLGA-NPs in aqueous phase.24,25 The colloidal nanosuspension of CIN-PLGA-NPs was centrifuged at 12,000 rpm for 30 min at 4°C to get the final NPs encapsulating CIN. CIN-PLGA-NPs were washed twice with deionized water to eliminate the unentrapped drug from the NPs surface. NPs were then re-dispersed in water.
Statistical Optimization of the Formulations
Experimental Design
We chose the amounts of 100 and 200 mg PLGA and the concentrations of 0.5% and 1% Poloxamer188 as the formulation parameters (independent variables, X1 and X2, respectively). Their effect on the CQAs of the developed formulations; entrapment efficiency (Y1), particle size (Y2), and zeta potential (Y3) was investigated using 2-factors, 2-levels design of experiment (Table 1). Thirteen formulations were fabricated based on a response surface central composite design (design-Expert R software version 12 state-ease, Inc., Minneapolis, MN, USA) as shown in Table 2.
 |
Table 1 The Experimental Range for Independent Variables and CQAs of CIN-PLGA-NPs |
 |
Table 2 Composition and the Observed Responses of the Design |
Characterization of CIN-PLGA-NPs
Entrapment Efficiency Determination (EE%)
CIN-PLGA-NPs were separated from dispersion by cooling centrifuge (2–16KL, Sigma Laborzentrifugen GmbH, Osterode am Harz, Germany) at 12,000 rpm for 2 hours at 4°C. After centrifugation, the obtained supernatant was diluted and filtered through 40-µm filter paper.26 After that, it was analyzed for free CIN spectrophotometrically using an ultraviolet-visible spectrophotometer (V-630, Jasco, Tokyo, Japan) at wavelength 270 nm against equivalent phosphate buffer as a blank.26,27 The EE % was calculated via the following equation:28
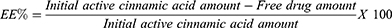
The given EE % values are the averages of three estimations.
Particle Size (PS), Polydispersity Index (PDI) and Zeta Potential (ZP) Measurement
PS, PDI and ZP of the prepared CIN-PLGA-NPs formulations were measured using a Zetasizer 3000 PCS (Malvern Instruments, England) equipped with a 5-mW helium-neon laser with a wavelength output of 633 nm. Measurements were accomplished at 25°C, angle 90°C, run time at least 180 sec. Interpretation of data was done by the method of cumulants. Prior to the measurements, the samples were diluted with deionized water. The PS, PDI and ZP values are the averages of three measurements.29
Design Space Optimization and Model Validation
The design space was created according to the desired criteria established for CQAs. The optimized formula was formulated and evaluated to compare its responses with the predicted values. Percentage error was calculated to validate the model. EE%, PS and ZP measurements of the optimized formulation were performed as previously mentioned.
Transmission Electron Microscopy (TEM)
The optimized CIN-PLGA-NPs formulation was diluted with deionized water, a drop of the sample was applied onto a mesh carbon-coated copper grid and allowed to settle for 35 min. Extra fluid was then eliminated by with the aid of an absorbent paper, then observed by TEM (JEM 1010, JEOL Ltd, Tokyo, Japan) with an acceleration voltage of 70kV and magnification power of 150 KX.29,30
In vitro Studies
In vitro Dissolution Studies
In vitro drug release of CIN from CIN-PLGA-NPs optimized formulation was performed utilizing the dialysis bag technique and compared with free CIN.31,32 Cellulose dialysis bag was presoaked in distilled water for 12 h. CIN-PLGA-NPs equivalent to 10 mg of CIN were exactly weighed and the same weight of free CIN was resuspended in phosphate buffer solution (PBS) of pH 7.4 and vortexed for 2 min. After that, they were transferred into the dialysis bags, and both ends of the bags were tightly closed. The bags were immersed into a 50 mL PBS of pH 7.4 which acted as the receptor cell, placed in a shaker water bath at 37±0.5 °C and shaked at 100 rpm. Aliquots were withdrawn from the receptor cell of each sample and substituted by equivalent volumes of the fresh release medium at time intervals from 1 to 24 h. Samples were analyzed spectrophotometrically at λmax 270 nm against PBS as a blank. Triplicate samples were measured and the average concentration was used. Additionally, in vitro drug release data of CIN from the prepared CIN-PLGA-NPs optimized formulation was investigated by fitting to various kinetic models and evaluated to explain the mechanism of drug release.
In vitro Cytotoxicity Assays
The triple-negative breast cancer cells, MDA-MB-231, were obtained from the American Type Culture Collection (ATCC, VA, USA). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) using 10% fetal bovine serum (FBS; Gibco USA) and 1% mixture of antimycotic/antibiotic (Gibco, USA). The cells were maintained under 5% CO2 and a temperature of 37 °C until they were ready for passaging. Cells were then seeded in 96-well plates at a density of 15×103 cells per well and kept in the incubator for 24 h. Treatment was then initiated using tenfold serial dilutions of either CIN-Free or CIN-PLGA-NPs at concentrations ranging from 0.002 to 20 mM for a period of 72 h. Viability was afterwards estimated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (SERVA, Germany), as previously described.33 Briefly, 20 µL of MTT reagent (5 mg/mL) was added to the wells and then left for 2 h. Dimethyl sulfoxide (DMSO) was then used to dissolve the formed formazan crystals. The optical density was the measured at 570 nm using a microplate reader (BioTek, USA). Viability (%) was then calculated from the absorbance detected in treated wells relative to that detected in the wells that received treatment vehicles only (control). Calculation of the half-maximal inhibitory concentration (IC50) was executed using GraphPad Prism software (GraphPad Software, v6, CA, USA).
Scratch Wound Assay
The migration potential of the triple-negative breast cancer cells, MDA-MB-231, with and without treatments was estimated using the scratch wound assay, as previously described.34 Briefly, MDA-MB-231 cells were seeded in 6-well plates at a density of 3×105 cells/well. When cells became confluent, the cell monolayer was scratched using a sterile pipette tip. Photomicrographs of the wounds at 0 h were taken. The cells were then incubated with either CIN-Free or CIN-PLGA-NPs and their vehicles serving as controls. Afterwards, cell migration into the scratch area was monitored over a period of time and photographed at 24 and 48 h under an inverted microscope equipped with a digital camera at 100× magnification. ImageJ software was then used to analyze the migration rate in the presence and absence of treatments by measuring the areas of the wounds (final area at 24 and 48 h) versus the original one (initial area at 0 h) to calculate the percentage of wound closure.
In vivo Experiments
Experimental Design
In vivo evaluation of the anticancer activity of the formulation was accomplished using an animal model of Ehrlich ascites carcinoma (EAC) solid tumor where Swiss albino female mice weighing between 20 and 25 gm were used. Approximately two million EAC cells/mouse were intramuscularly inoculated after collecting them from an ascitic-tumor-bearing mouse. Once tumor sizes reached an average of 200 mm3, treatments were initiated. Thirty female mice were divided into the following groups (n=10): control group where mice received drug vehicles only, CIN-Free group where mice received 250 mg/kg of free CIN, i.p., 3 times/week for 21 days, and CIN-PLGA-NPs group where mice were given an i.p. injection of CIN-PLGA-NPs equivalent to 250 mg/kg of free CIN, 3 times/week for 21 days. Six days after inoculation of EAC cells, treatments were initiated for a period of 21 days. Body weights were also periodically recorded for all mice belonging to different groups. Moreover, mice were monitored for acute signs of toxicity for evaluation of the safety profile of the used formulation. Mice were sacrificed under anesthesia and tumors were collected for further analysis. Experiments were carried out in accordance with the Guide for Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85–23, revised 2011) and were approved by the Ethics Committee of the Faculty of Pharmacy, The British University in Egypt.
Tumor Volume
Estimation of tumor volumes was achieved using a digital caliper at 0, 3, 6, 9, 12, 15, 18, 21 days post treatment initiation, by applying the following equation35:
“L” is the tumor length and “W” is the tumor width.
Histopathological Examination and Determination of Necrotic and Apoptotic Indices
After excision of tumors from mice, tissues were fixed in formalin solution (10%) and further embedded in paraffin. Then, 4 µm-thick sections were prepared and stained using hematoxylin and eosin (H&E) for investigation at two magnification powers (×100 and ×400) using a light microscope (Leica, Germany) in order to determine the histopathological changes in control and treated groups.
Necrotic and apoptotic indices were estimated by detecting the necrotic and apoptotic percentages in 10 random fields per section for each mouse at ×100 magnification power.
Estimation of Caspase-3 Immunoreactivity in Solid Tumors
An avidin-biotin complex immunoperoxidase technique was executed for immunohistochemical staining of tumor sections. A monoclonal antibody against caspase-3 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used after dilution (1:100). For detection, streptavidin-biotin-peroxidase complex and peroxidase- 3,3′-diaminobenzidine (DAB) were used, according to the manufacturer’s instructions (Dako, Denmark). Mayer’s hematoxylin was used for further counterstaining. Expression was then semi-quantitatively estimated as percentage of positively stained cells in 10 random fields per each section per mouse.36
Estimation of E-Cadherin and N-Cadherin Levels in Solid Tumors
Tumor tissue homogenates were prepared in 10% PBS. Then, protein content was quantified using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, USA). E- and N-cadherin levels in tumor homogenates were then estimated using E-cadherin and N-cadherin mouse ELISA kits (MyBioSource, USA), as per the manufacturers’ instructions. Absorbance was measured at 450 nm using a microplate reader.
Statistical Analysis
Data are illustrated as mean ± SD and were analyzed by one-way ANOVA followed by Tukey Kramer’s post hoc test for multiple comparisons using GraphPad Prism software (v6, CA, USA). Significance was counted at P<0.05.
Results and Discussion
Response Surface Central Composite Design and Analysis
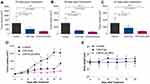
Thirteen runs were developed in accordance with the experimental design. Formulations’ composition in addition to the observed responses of the design are displayed in Table 2. It was noticed that the predicted R2 values were in agreement with the adjusted R2 values in all studied responses (within approximately 0.2 of each other). The adequate precision value is for confirming the model adequacy to navigate the design space in which a ratio more than 4 is desirable, and that was observed in all examined responses. The impacts of the independent variables on the dependent ones were further elucidated using three-dimensional response surface plots and contour plots (Figure 1). Figures S1–S13 in the supplementary materials show the PS distribution and ZP of all the prepared formulations.
 |
Figure 1 Response 3D plots (A, B and C) and Contour plots (D, E and F) for the effect of PLGA amount (X1) and Poloxamer188 concentration (X2) on EE%, PS and ZP. |
Impact of Formulation Factors on CQAs
EE %
EE% of CIN-PLGA-NPs formulations ranged from 60.13±0.20% to 80.19±1.29%, as presented in Table 2. The data were analyzed using polynomial analysis fitted with quadratic model with P<0.0001 indicating significance of the model.
The predicted and adjusted R2 were 0.8888 and 0.9470, respectively, indicating good correlation. The lack of fit (P=0.5676) was not significant revealing that the equation was a good fit and the adequate precision was 20.88. The polynomial equation that reveals the relation between EE % and independent formulation parameters is as follows:
From the quadratic equation and Figure 1A and D, it is clear that the EE % is positively affected by PLGA amount (X1) and negatively affected by Poloxamer188 concentration (X2). The increase in X1 significantly increased the EE% at P< 0.0001 and this could be attributed to the fact that higher concentration of polymer provides more viscous organic solution and hence lower net shear stress, which would result in particles with larger size and sufficient surface for CIN to be entrapped.37 However, increasing X2 significantly reduced EE% (P=0.0011) which may be due to the solubilization of CIN in excess Poloxamer188.38
PS
PS of the prepared CIN-PLGA-NPs formulations ranged from 144.9±9.63 nm to 260.3±20.17 nm as illustrated in Table 2. Data obtained were analyzed by applying polynomial analysis fitted with quadratic model with P<0.0001 indicating significance of the model. The predicted R2 (0.9079) and the adjusted R2 (0.9724) were in good correlation. The Lack of F (P=0.1072) was not significant and the adequate precision was 28.54.
The quadratic equation demonstrating the relation between PS and independent formulation parameters is as follows:
From Figure 1B and E and the quadratic equation, it can observed that increasing X1 significantly increased the PS (P<0.0001) and this result greatly matched with the already available literature studies.21 On the other hand, we observed that the mean size of CIN-PLGA-NPs decreased with increasing Poloxamer188 concentration. As a surfactant, Poloxamer188 employs its stabilizing effect by being adsorbed at the NPs interface, hence reducing the surface tension between the two phases, avoiding the NPs aggregation, thereby decreasing their size.39
Regarding PDI, an indicator of the size distribution homogeneity,40 NPs displayed relatively monodisperse size distributions with a PDI below 0.2.41 The PDI values of the prepared formulations ranged from 0.047±0.072 to 0.245±0.089 indicating that CIN-PLGA-NPs are stable NPs with a relatively narrow size distribution.42 PDI values were in the acceptable desired range required for further studies.
ZP
ZP is an important physicochemical parameter that influences stability of the NPs. ZP of CIN-PLGA-NPs formulations ranged from −19.1±1.66 to −32.9±0.70 mV, as shown in Table 2. The CIN-PLGA-NPs fabricated formulations had a negative electric charge, which could be attributed to the ionized terminal carboxylic groups of PLGA present on the surface of the NPs.37 The electrostatic repulsion between NPs with the same electric charge prevents their aggregation.43 ZP values of −15 to −30 mV are common for well-stabilized NPs.44 Therefore, it was concluded that the prepared formulations would remain stable under storage.
The linear model was the most appropriate for analyzing data with P<0.0001 indicating the significance of the model. The predicted R2 was 0.8560 and the adjusted R2 was 0.9006 representing good correlation. The Lack of F (P=0.2622) was not significant, and the adequate precision was 21.71.
The equation demonstrating the relation between ZP and independent formulation factors is as follows:
From Figure 1C and F and the linear equation, it can be concluded that increasing X1 significantly increased the ZP (P<0.0001) which is in agreement with the findings of Anwer et al.42 Additionally, increasing X2 has significantly decreased the ZP (P< 0.0001). This may be attributed to the decreasing effect of the Poloxamer188 on the particles’ electrostatic repulsion.40
Selection of the Optimized CIN-PLGA-NPs
An optimum formulation was achieved after response surface analysis of the experimental factors using central composite design based on the conditions for attaining maximum EE% and ZP in addition to minimum PS. The CIN-PLGA-NPs optimized formula was developed by using PLGA amount of 161.9 mg (X1) and Poloxamer188 concentration of 0.5% (X2). The optimized formula predicted to have EE% of 76.98%, PS of 186.3 nm and ZP of −28.47 mV. In addition, it was evaluated and compared to the predicted values and exhibited a percentage error of −0.5, 2.1 and 0.36 for EE%, PS and ZP, respectively. This low magnitude of error indicates that central composite model is adequate and provides high prediction in optimizing CIN-PLGA-NPs formulations. Moreover, the optimized formula showed PDI of 0.070 suggesting reasonable homogeneity of the formulation.
Transmission Electron Microscopy (TEM)
Morphological analysis of the selected formula is shown in Figure 2. The CIN-PLGA-NPs optimized formula appeared in a smooth spherical shape with relatively uniform size distribution, as observed by TEM micrographs. The size of CIN-PLGA-NPs obtained from TEM examination was in agreement with the PS analysis.
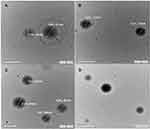 |
Figure 2 Transmission electron micrographs of CIN-PLGA-NPs with magnification of 20 kx (A and B), 15kx (C) and 8 kx (D). |
In vitro Studies
In vitro Dissolution Studies
The in vitro dissolution profile of CIN-PLGA-NPs and CIN-Free is shown in Figure 3. The release profile demonstrated an initial burst effect of CIN from CIN-PLGA-NPs in the first 4 hours (25%), which can be attributed to a large amount of CIN that was close to or attached to the surface of the particles.45 After the initial burst release, CIN-PLGA-NPs showed a sustained release behavior which may be influenced by the rate at which PLGA degrades by hydrolysis of the nanoparticle skeleton.46 The burst release of CIN from CIN-PLGA-NPs may also be due to the dissolution and diffusion of the CIN that was poorly entrapped in the PLGA matrix, whereas the slower and continuous release was likely due to the diffusion of the CIN localized in the PLGA core of the CIN-PLGA-NPs. These findings are in line with a relevant study executed using azithromycin PLGA nanoparticles.47 It was also found that the dissolution rate of CIN-PLGA-NPs was faster than those of CIN-Free. The obtained data showed that the optimized formulation reached 61% cumulative release after 24 h, in comparison to 27% only of the CIN-Free. This could be attributed to the small PS of the NPs that led to increase in the surface area, and hence enhanced dissolution rate.13 Moreover, the solubility of the CIN-Free can also be improved by reducing the PS according to Oswald-Freundlich equation.14 Therefore, the CIN-PLGA-NPs had the ability to improve the bioavailability owing to the enhanced solubility and dissolution rate.48 The in vitro CIN release from CIN-PLGA-NPs was examined on several kinetic models (zero-order, first-order, and Higuchi equations), to determine the drug release mechanism. The highest r value for drug release was attained from Higuchi diffusion’s equation at 0.967637 compared to that of zero-order at 0.909122 and first order at 0.9574. Accordingly, the mechanism of drug release is thought to be diffusion. Additionally, we concluded that the release of CIN from CIN-PLGA-NPs follows a sustained release pattern.
 |
Figure 3 In-vitro dissolution profile of CIN-PLGA-NPs compared to CIN-Free. |
Effect of CIN-PLGA-NPs on the Cytotoxicity of Triple-Negative Breast Cancer Cells
CIN was previously found to have cytotoxic effect in different cancer cell types including breast cancer.6 In addition, the anti-proliferative potential of CIN on MDA-MB-231 cell line was previously reported.49 Accordingly, the present study aimed to compare the cytotoxic effect of CIN-PLGA-NPs to that of CIN-Free on triple negative MDA-MB-231 cell line hypothesizing that nanoencapsulation would enhance the biological effect of the free drug. As shown in Figure 4, CIN-PLGA-NPs exhibited a cytotoxic potential on the triple-negative breast cancer cell line, MDA-MB-231, where the reported IC50 was 0.5171 mM versus 2.296 mM in the cells treated with free CIN. These results imply that drug encapsulation by NPs could improve the cytotoxicity of free drugs. The potential cytotoxic effect of CIN-PLGA-NPs over CIN-Free could be due to the enhanced drug solubility and small PS that allows better cellular uptake and cell internalization, leading to intracellular release of CIN, and therefore, improved anticancer activity. It is worth mentioning that previous reports shed light on the anticancer activity of CIN and its derivatives in many types of cancer such as glioblastoma and prostate cancer.50 In addition, the study of Pal et al49 provided mechanistic insights to elucidate the underlying anticancer activity of CIN in triple-negative breast cancer reporting the induction of an extrinsic apoptotic pathway.
 |
Figure 4 Effect of CIN-Free and CIN-PLGA-NPs on the viability of MDA-MB-231 breast cancer cell lines, as detected by MTT assay. |
Effect of CIN-PLGA-NPs on Migration of MDA-MB-231 Cells
To investigate the effect of CIN-PLGA-NPs on the migratory potential of metastatic MDA-MB- 231 cells, the scratch wound healing assay was executed. As shown in Figure 5A–C, increases in wound closure percentage were reported with CIN-PLGA-NPs relative to the control and CIN-Free-treated cells at 48 h reaching 6.68 (P<0.001) and 1.8 (P=0.008) folds, respectively, whereas at 24 h, only the CIN-PLGA-NPs were capable of increasing the wound closure significantly by 1.65 folds (P=0.0042), as compared to the control untreated cells. Although, trans-cinnamic acid was previously reported to induce fibroblast migration,51 its impact on triple-negative breast cancer cells migratory potential remains heretofore unexplored. Moreover, albeit CIN-Free did not fully curtail MDA-MB-231 cell migration as evidenced by the wound closure results, CIN-PLGA-NPs demonstrated higher potential in this aspect. This could be attributed to the small PS of the CIN-PLGA-NPs that could have enhanced drug solubility and cell uptake, thereby achieving better impact on cell migration over the free form. Among the limitations of the in vitro experiments in the present study is the lack of further molecular assays confirming the inhibitory potential of the CIN-PLGA-NPs on the migration triple-negative breast cancer cells.
In vivo Experiments
Effect of CIN-PLGA-NPs on Tumor Volumes
Tumor volumes were measured at different time points throughout the experiment (days 0, 3, 6, 9, 12, 15, 18, and 21) in tumor-bearing mice. As shown in Figure 6A–D, CIN-PLGA-NPs were capable of decreasing tumor volumes at earlier time points of the experiment compared to CIN-Free where significant changes were reported in the CIN-PLGA-NPs group starting the ninth day after treatment initiation with no significant changes reported in the CIN-Free-treated group versus positive control at the same time point. At the 15th, 18th and 21st days post treatment, changes were more conspicuous where CIN-Free-treated group showed reductions in tumor volumes reaching 55.7% (P=0.0282), 68% (P=0.0005) and 45% (P=0.033), respectively, as compared to the control untreated group. Meanwhile, CIN-PLGA-NPs demonstrated reduced tumor volumes reaching 77% (P = 0.002), 80.4% (P <0.0001) and 75.6% (P =0.005) compared to the control untreated mice group at the 15th, 18th, and 21st days post treatment, respectively. The study of Almeer et al52 corroborated the present findings where a reduction in tumor volume was previously reported with CIN in an EAC solid tumor animal model. However, CIN-PLGA-NPs, as presented herein demonstrated superior capacity in reducing tumor size over CIN-Free. These findings are in agreement with the in vitro cytotoxicity assays’ results.
Evaluation of CIN-PLGA-NPs Overall Safety Profile
Body weights were recorded at different time points throughout the experiment (0, 4, 8, 12, 16, and 20 days after treatment). Notably, no changes were reported in body weights among different animal groups (Figure 6E). Moreover, the animal’s excitability and vitality were observed over the course of the experiment where no changes were reported in the aforementioned aspects. Also, no short-term acute signs of toxicity were observed. Additionally, no mortality was reported in the present study in the different groups. These findings are therefore indicative of the CIN-PLGA-NPs formulation safety. However, investigating the integrity of the most perfused organs such as the liver, spleen, heart, and kidneys is recommended to further confirm the safety profile of the newly developed formulation.
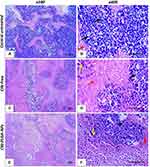
Effect of CIN-PLGA-NPs on Histopathological Findings in Tumor Sections
As shown in Figure 7A and B, histopathological examination of tumor samples showed intact malignant cells, small and large higher chromatophilic tumor cells of variable shape representing cell proliferation in the control untreated group and areas of necrosis and apoptosis less than 5%. In the CIN-Free-treated group (Figure 7C and D), moderate number of malignant cells showing moderate necrosis, apoptosis, apoptotic bodies, karyorrhexis and/or karyolysis of nuclei were detected in the necrotic areas. On the other hand, CIN-PLGA-NPs-treated group, demonstrated fewer number of malignant cells with marked apoptosis as well as necrosis, apoptotic bodies, karyorrhexis and/or karyolysis of nuclei in necrotic areas (Figure 7E and F).
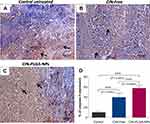
Effect of CIN-PLGA-NPs on Caspase-3 Immunoreactivity in Tumor Sections
Among all caspases, caspase-3, an executioner caspase, is considered the mainstay of the apoptogenic process in breast cancer. Cleaved caspase-3, which represents its active form, instigates changes in functional proteins within the cell thereby driving cancer cells towards apoptosis.53 In the current study, the immunohistochemical expression of caspase-3 was investigated in tumor sections to reflect on the pro-apoptotic potential of the optimized formulation. Our findings showed an elevation in caspase-3 immunoreactivity in tumor sections affiliated to both CIN-free and CIN-PLGA-NPs groups. Interestingly, the group which received CIN-PLGA-NPs exhibited significantly higher caspase-3 expression, as compared to the CIN-free-treated group, by 1.5 fold (P=0.0009) (Figure 8). The recent study of Pal et al49 revealed the capability of cinnamic acid to induce apoptosis in triple-negative breast cancer by enhancing caspase-3 levels in MDA-MB-231 cells. The present findings also insinuate a superior potential of CIN-PLGA-NPs over the free form in inducing apoptosis via caspase-3 enhancement.
Effect of CIN-PLGA-NPs on Necrotic and Apoptotic Indices in Tumor Sections
Necrotic and apoptotic indices were semi-quantitatively estimated in tumor sections stained with H&E. As shown in Figure 9A, the necrotic indices increased by 4.7 (P=0.0189) and 8.5 (P=0.0005) folds in CIN-Free and CIN-PLGA-NPs groups, respectively, as compared to the control untreated group. Interestingly, the group which received CIN-PLGA-NPs demonstrated an increased necrotic index compared to CIN-Free by 1.8 fold (P=0.0164). Moreover, the apoptotic index in the CIN-Free- and CIN-PLGA-NPs-treated groups where higher by 5.7 (P=0.0032) and 11 (P<0.0001) folds, as compared to the control untreated group, respectively. It is noteworthy that CIN-PLGA-NPs exhibited a higher apoptotic index by 2 folds as compared to the positive control untreated group (P=0.0015) (Figure 9B). Induction of apoptosis via influencing caspase-3, Bax, and Bcl-xl by CIN was reported in previous studies.50
Effect of CIN-PLGA-NPs on E-Cadherin and N-Cadherin Levels in Solid Tumors
Phenotypic changes are considered hallmarks for cells undergoing EMT. These cells display decreased expression of the epithelial marker, E-cadherin, concomitant with an increased expression of the mesenchymal marker, N-cadherin.54 The tumor levels of both E-cadherin and N-cadherin were estimated in the present study to reflect on the EMT status. Although the effect of cinnamic acid on EMT was not amply explored in cancer, few reports disclosed a likely role of cinnamic acid in the healing process by improving fibroblasts migration.51 However, its impact – apart from its derivatives – on migration of cancer cells was not hitherto investigated. The current study, therefore, offers insights on how cinnamic acid might influence migration in a cancer setting, shedding light on E- and N-cadherin levels in tumor tissues to stand on both epithelial and mesenchymal states. As shown in Figure 9C, CIN-PLGA-NPs caused an increase in E-cadherin levels by 1.2 fold compared to the control untreated group. Meanwhile, tumor levels of N-cadherin were reduced in the CIN-PLGA-NPs-treated group by 30%, as compared to the CIN-Free-treated group, whereas the group that received free CIN failed to show any significant change in either E- or N-cadherin at the specified dose (Figure 9D). Accordingly, we can conclude that a likely impact on EMT exerted by the CIN-PLGA-NPs exists which might have been reflected on the metastatic potential of cancer cells. These findings, however, need further studies using a xenograft or a syngeneic animal model for triple-negative breast cancer to confirm.
Conclusion
In the current study, a novel CIN-PLGA-NPs delivery system was successfully developed by nanoprecipitation method to improve the biological effects of CIN on triple-negative breast cancer. The loading of CIN into PLGA nanoparticles led to enhanced solubility, dissolution rate, bioavailability and hence improved biological efficacy. Regarding the impact on the pro-apoptotic and migratory potentials, the prepared optimized formulation showed significant superiority in both in vitro and in vivo studies over CIN-Free. Moreover, the present study provides preliminary data on the overall safety profile of the optimized formula, that would endow eligibility for further pre-clinical and clinical future investigations. Overall, our findings suggest the use of the optimized CIN-PLGA-NPs in triple-negative breast cancer patients to improve the therapeutic outcomes and prevent metastasis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sun Y-S, Zhao Z, Yang Z-N, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387. doi:10.7150/ijbs.21635
2. El-Kersh DM, Ezzat SM, Salama MM, et al. Anti-estrogenic and anti-aromatase activities of citrus peels major compounds in breast cancer. Sci Rep. 2021;11(1):1–14. doi:10.1038/s41598-021-86599-z
3. Chowdhury P, Ghosh U, Samanta K, Jaggi M, Chauhan SC, Yallapu MM. Bioactive nanotherapeutic trends to combat triple negative breast cancer. Bioact Mater. 2021;6(10):3269–3287. doi:10.1016/j.bioactmat.2021.02.037
4. Yuan Y, Cai T, Xia X, Zhang R, Chiba P, Cai Y. Nanoparticle delivery of anticancer drugs overcomes multidrug resistance in breast cancer. Drug Deliv. 2016;23(9):3350–3357. doi:10.1080/10717544.2016.1178825
5. Wang S, Zhou Y, Tao W, Liu J, Chen H, Zhao Z. Fe3O4-modified amphiphilic polyurethane nanoparticles with good stability as magnetic-targeted drug carriers. Polym Bull. 2021:1–15. doi:10.1007/s00289-021-03931-3
6. Hunke M, Martinez W, Kashyap A, Bokoskie T, Pattabiraman M, Chandra S. Antineoplastic actions of cinnamic acids and their dimers in breast cancer cells: a comparative study. Anticancer Res. 2018;38(8):4469–4474. doi:10.21873/anticanres.12749
7. Khaled N, Bidet Y. New insights into the implication of epigenetic alterations in the EMT of triple negative breast cancer. Cancers. 2019;11(4):559. doi:10.3390/cancers11040559
8. Xu X, Zhang L, He X, et al. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun. 2018;502(1):160–165. doi:10.1016/j.bbrc.2018.05.139
9. Du B, Shim JS. Targeting epithelial–mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21(7):965. doi:10.3390/molecules21070965
10. Ruwizhi N, Aderibigbe BA. Cinnamic acid derivatives and their biological efficacy. Int J Mol Sci. 2020;21(16):5712. doi:10.3390/ijms21165712
11. Letsididi KS, Lou Z, Letsididi R, Mohammed K, Maguy BL. Antimicrobial and antibiofilm effects of trans-cinnamic acid nanoemulsion and its potential application on lettuce. LWT. 2018;94:25–32. doi:10.1016/j.lwt.2018.04.018
12. Martínez-Rosas JR, Díaz-Torres R, Ramírez-Noguera P, López-Barrera LD, Escobar-Chavez JJ, Ángeles ER. PLGA nanoparticles of a new cinnamic acid derivative inhibits cellular proliferation on breast cancer cell line MCF-7 in a PPARγ dependent way. Die Pharmazie. 2020;75(7):324–328.
13. Li W, Zhao X, Sun X, Zu Y, Liu Y, Ge Y. Evaluation of antioxidant ability in vitro and bioavailability of trans-cinnamic acid nanoparticle by liquid antisolvent precipitate. J Nanomater. 2016;2016:1–11. doi:10.1155/2016/9518362
14. Sathya S, Shanmuganathan B, Saranya S, Vaidevi S, Ruckmani K, Pandima Devi K. Phytol-loaded PLGA nanoparticle as a modulator of Alzheimer’s toxic Aβ peptide aggregation and fibrillation associated with impaired neuronal cell function. Artif Cells Nanomed Biotechnol. 2018;46(8):1719–1730. doi:10.1080/21691401.2017.1391822
15. Al-Jubori AA, Sulaiman GM, Tawfeeq AT, Mohammed HA, Khan RA, Mohammed SA. Layer-by-layer nanoparticles of tamoxifen and resveratrol for dual drug delivery system and potential triple-negative breast cancer treatment. Pharmaceutics. 2021;13(7):1098. doi:10.3390/pharmaceutics13071098
16. Brigger I, Dubernet C, Couvreur P. Nanoparticles in cancer therapy and diagnosis. Adv Drug Deliv Rev. 2012;64:24–36. doi:10.1016/j.addr.2012.09.006
17. Haggag YA, Abosalha AK, Tambuwala MM, et al. Polymeric nanoencapsulation of zaleplon into PLGA nanoparticles for enhanced pharmacokinetics and pharmacological activity. Biopharm Drug Dispos. 2021;42(1):12–23. doi:10.1002/bdd.2255
18. Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41(7):2971–3010. doi:10.1039/c2cs15344k
19. De Vita A, Liverani C, Molinaro R, et al. Lysyl oxidase engineered lipid nanovesicles for the treatment of triple negative breast cancer. Sci Rep. 2021;11(1):1–12. doi:10.1038/s41598-021-84492-3
20. Zou T, Lan M, Liu F, et al. Emodin-loaded polymer-lipid hybrid nanoparticles enhance the sensitivity of breast cancer to doxorubicin by inhibiting epithelial–mesenchymal transition. Cancer Nanotechnol. 2021;12(1):1–15. doi:10.1186/s12645-021-00093-9
21. Ahmad N, Alam MA, Ahmad R, Naqvi AA, Ahmad FJ Preparation and characterisation of surface-modified PLGA-polymeric nanoparticles used to target treatment of intestinal cancer (Retraction of Vol 46, Pg 432, 2018). Abingdon, Oxon: Taylor & Francis Ltd; 2020.
22. Li Z, Jiang H, Xu C, Gu L. A review: using nanoparticles to enhance absorption and bioavailability of phenolic phytochemicals. Food Hydrocoll. 2015;43:153–164. doi:10.1016/j.foodhyd.2014.05.010
23. Shirsat AE, Chitlange SS. Application of quality by design approach to optimize process and formulation parameters of rizatriptan loaded chitosan nanoparticles. J Adv Pharm Technol Res. 2015;6(3):88. doi:10.4103/2231-4040.157983
24. Budhian A, Siegel SJ, Winey KI. Haloperidol-loaded PLGA nanoparticles: systematic study of particle size and drug content. Int J Pharm. 2007;336(2):367–375. doi:10.1016/j.ijpharm.2006.11.061
25. Lancheros R, Guerrero CA, Godoy-Silva RD. Improvement of N-acetylcysteine loaded in PLGA nanoparticles by nanoprecipitation method. J Nanotechnol. 2018;2018:1–11. doi:10.1155/2018/3620373
26. Sah AK, Suresh PK, Verma VK. PLGA nanoparticles for ocular delivery of loteprednol etabonate: a corneal penetration study. Artif Cells, Nanomed Biotechnol. 2017;45(6):1156–1164. doi:10.1080/21691401.2016.1203794
27. Zhang Y-T, Xu Y-M, Zhang S-J, et al. In vivo microdialysis for the evaluation of transfersomes as a novel transdermal delivery vehicle for cinnamic acid. Drug Dev Ind Pharm. 2014;40(3):301–307. doi:10.3109/03639045.2012.756888
28. Pandit J, Sultana Y, Aqil M. Chitosan-coated PLGA nanoparticles of bevacizumab as novel drug delivery to target retina: optimization, characterization, and in vitro toxicity evaluation. Artif Cells Nanomed Biotechnol. 2017;45(7):1397–1407. doi:10.1080/21691401.2016.1243545
29. Khalifa MK, Salem HA, Shawky SM, Eassa HA, Elaidy AM. Enhancement of zaleplon oral bioavailability using optimized self-nano emulsifying drug delivery systems and its effect on sleep quality among a sample of psychiatric patients. Drug Deliv. 2019;26(1):1243–1253. doi:10.1080/10717544.2019.1687613
30. Panda BP, Krishnamoorthy R, Shivashekaregowda NKH, Patnaik S. Influence of Poloxamer-188 on design and development of second generation PLGA nanocrystals of metformin hydrochloride. Nano Biomed Eng. 2018;10(4):334–343. doi:10.5101/nbe.v10i4.p334-343
31. Yang SC, Lu LF, Cai Y, Zhu JB, Liang BW, Yang CZ. Body distribution in mice of intravenously injected camptothecin solid lipid nanoparticles and targeting effect on brain. J Control Release. 1999;59(3):299–307. doi:10.1016/S0168-3659(99)00007-3
32. Nasr AM, Elhady SS, Swidan SA, Badawi NM. Celecoxib loaded in-situ provesicular powder and its in-vitro cytotoxic effect for cancer therapy: fabrication, characterization, optimization and pharmacokinetic evaluation. Pharmaceutics. 2020;12(12):1157. doi:10.3390/pharmaceutics12121157
33. Mosdam T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxic assay. J Immunol Methods. 1983;65(1–2):55–63. doi:10.1016/0022-1759(83)90303-4
34. Liang -C-C, Park AY, Guan J-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329–333. doi:10.1038/nprot.2007.30
35. Jaganathan SK, Mondhe D, Wani ZA, Pal HC, Mandal M. Effect of honey and eugenol on Ehrlich ascites and solid carcinoma. J Biomed Biotechnol. 2010;2010:1–5. doi:10.1155/2010/989163
36. Hsu S-M, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29(4):577–580. doi:10.1177/29.4.6166661
37. Tefas LR, Tomuţă I, Achim M, Vlase L. Development and optimization of quercetin-loaded PLGA nanoparticles by experimental design. Clujul Med. 2015;88(2):214. doi:10.15386/cjmed-418
38. Bian X, Liang S, John J, et al. Development of PLGA-based itraconazole injectable nanospheres for sustained release. Int J Nanomedicine. 2013;8:4521. doi:10.2147/IJN.S54040
39. Jonderian A, Maalouf R. Formulation and in vitro interaction of rhodamine-B loaded PLGA nanoparticles with cardiac myocytes. Front Pharmacol. 2016;7:458. doi:10.3389/fphar.2016.00458
40. Schwarz C, Mehnert W, Lucks JS, Müller RH. Solid lipid nanoparticles (SLN) for controlled drug delivery. I. Production, characterization and sterilization. J Control Release. 1994;30(1):83–96. doi:10.1016/0168-3659(94)90047-7
41. García-Díaz M, Foged C, Nielsen HM. Improved insulin loading in poly (lactic-co-glycolic) acid (PLGA) nanoparticles upon self-assembly with lipids. Int J Pharm. 2015;482(1–2):84–91. doi:10.1016/j.ijpharm.2014.11.047
42. Anwer MK, Mohammad M, Ezzeldin E, Fatima F, Alalaiwe A, Iqbal M. Preparation of sustained release apremilast-loaded PLGA nanoparticles: in vitro characterization and in vivo pharmacokinetic study in rats. Int J Nanomedicine. 2019;14:1587. doi:10.2147/IJN.S195048
43. Öztürk AA, Martin-Banderas L, Cayero-Otero MD, Yenilmez E, Yazan Y. New approach to hypertension treatment: carvediol-loaded PLGA nanoparticles, preparation, in vitro characterization and gastrointestinal stability. Lat Am J Pharm. 2018;37(9):1730–1741.
44. Kesisoglou F, Panmai S, Wu Y. Nanosizing—oral formulation development and biopharmaceutical evaluation. Adv Drug Deliv Rev. 2007;59(7):631–644. doi:10.1016/j.addr.2007.05.003
45. Sansdrap P, Moës AJ. In vitro evaluation of the hydrolytic degradation of dispersed and aggregated poly (DL-lactide-co-glycolide) microspheres. J Control Release. 1997;43(1):47–58. doi:10.1016/S0168-3659(96)01469-1
46. Wang L, Wang Y, Wang X, et al. Encapsulation of low lipophilic and slightly water-soluble dihydroartemisinin in PLGA nanoparticles with phospholipid to enhance encapsulation efficiency and in vitro bioactivity. J Microencapsul. 2016;33(1):43–52. doi:10.3109/02652048.2015.1114042
47. Mohammadi G, Valizadeh H, Barzegar-Jalali M, et al. Development of azithromycin–PLGA nanoparticles: physicochemical characterization and antibacterial effect against Salmonella typhi. Colloids Surf B Biointerfaces. 2010;80(1):34–39. doi:10.1016/j.colsurfb.2010.05.027
48. Wang H, Li Q, Deng W, et al. Self‐nanoemulsifying drug delivery system of trans‐cinnamic acid: formulation development and pharmacodynamic evaluation in alloxan‐induced type 2 diabetic rat model. Drug Dev Res. 2015;76(2):82–93. doi:10.1002/ddr.21244
49. Pal A, Tapadar P, Pal R. Exploring the molecular mechanism of cinnamic acid-mediated cytotoxicity in triple negative MDA-MB-231 breast cancer cells. Curr Med Chem Anticancer Agents. 2021;21(9):1141–1150.
50. Imai M, Yokoe H, Tsubuki M, Takahashi N. Growth inhibition of human breast and prostate cancer cells by cinnamic acid derivatives and their mechanism of action. Biol Pharm Bull. 2019:b18–01002. doi:10.1248/bpb.b18-01002
51. De Aquino FLT, Da Silva JP, de Souza Ferro JN, Lagente V, Barreto E. Trans-Cinnamic acid, but not p-coumaric acid or methyl cinnamate, induces fibroblast migration through PKA-and p38-MAPK signalling pathways. J Tissue Viability. 2021;30:363–371. doi:10.1016/j.jtv.2021.05.003
52. Almeer RS, Aref AM, Hussein RA, Othman MS, Abdel Moneim AE. Antitumor potential of berberine and cinnamic acid against solid Ehrlich carcinoma in mice. Curr Med Chem Anticancer Agents. 2019;19(3):356–364.
53. Pu X, Storr SJ, Zhang Y, et al. Caspase-3 and caspase-8 expression in breast cancer: caspase-3 is associated with survival. Apoptosis. 2017;22(3):357–368. doi:10.1007/s10495-016-1323-5
54. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi:10.1038/nrm3758
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.