Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
Invasive Breast Cancer with HER2 ≥4.0 and <6.0: Risk Classification and Molecular Typing by a 21-Gene Expression Assay and MammaPrint Plus BluePrint Testing
Authors Bai Q , Lv H , Bao L, Yang Y, Zhang X, Chang H, Xue T, Ren M, Zhu X, Zhou X, Yang W
Received 24 May 2023
Accepted for publication 26 July 2023
Published 3 August 2023 Volume 2023:15 Pages 563—575
DOI https://doi.org/10.2147/BCTT.S420738
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Robert Clarke
Qianming Bai,1– 3,* Hong Lv,1– 3,* Longlong Bao,1– 3,* Yu Yang,1– 3 Xin Zhang,4 Heng Chang,1– 3 Tian Xue,1– 3 Min Ren,1– 3 Xiaoli Zhu,1– 3 Xiaoyan Zhou,1– 3,* Wentao Yang1– 3,*
1Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, 200032, People’s Republic of China; 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 200032, People’s Republic of China; 3Institute of Pathology, Fudan University, Shanghai, 200032, People’s Republic of China; 4Department of Pathology, Fudan University Zhongshan Hospital, Shanghai, 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wentao Yang; Xiaoyan Zhou, Department of Pathology, Fudan University Shanghai Cancer Centre, 270 Dong’an Road, Shanghai, 200032, People’s Republic of China, Tel +86-18017312353 ; +86-18017312385, Email [email protected]; [email protected]
Purpose: To investigate the HER2 status and clinicopathological features in invasive breast cancer with HER2 ≥ 4.0 and < 6.0, which has always been controversial.
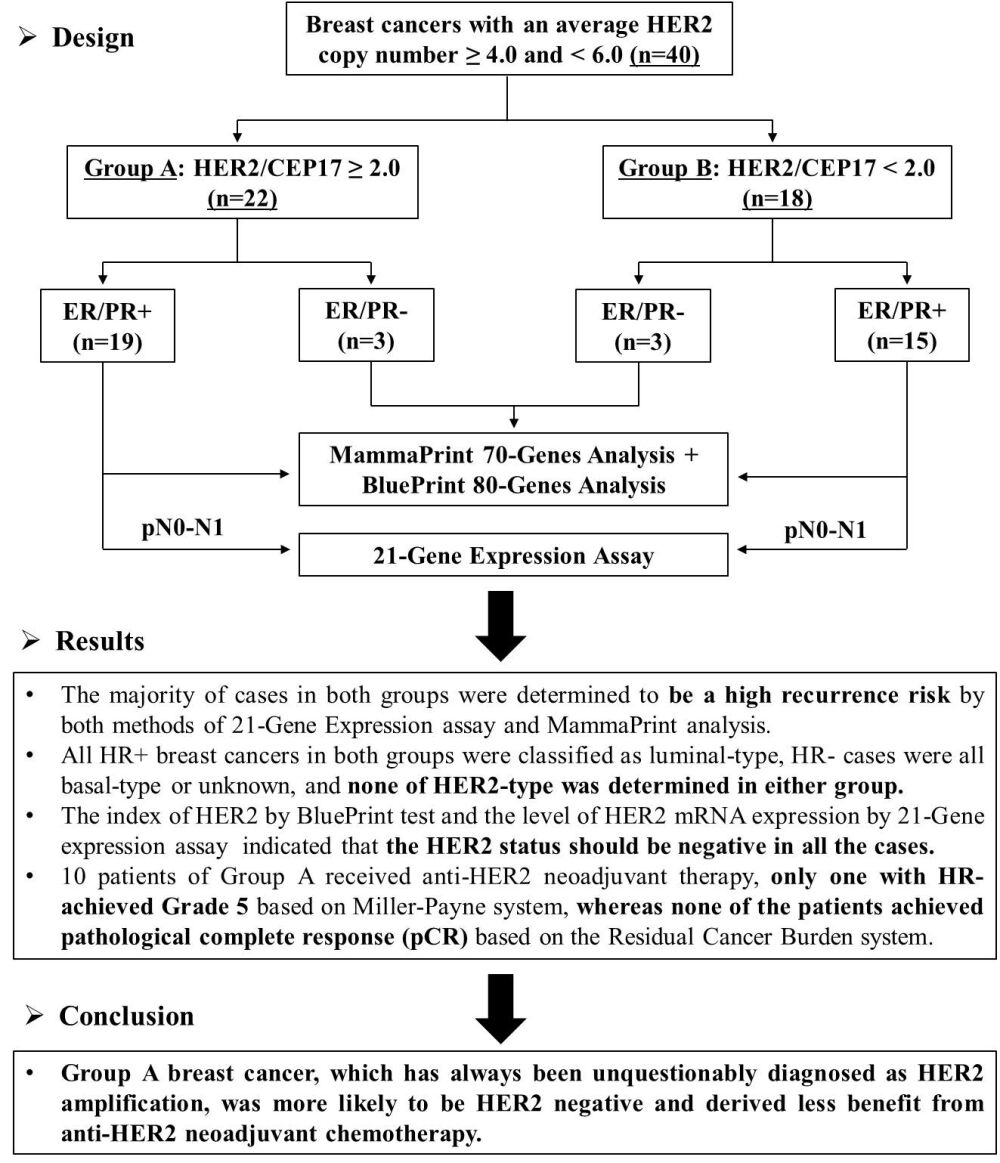
Methods: Forty breast cancer cases with HER2 ≥ 4.0 and < 6.0 by fluorescence in situ hybridization (FISH) were collected and classified into two groups based on the HRE2/CEP17 ratio (Group A: ≥ 2.0, n=22; Group B: < 2.0, n=18). Clinicopathological characteristics, HER2 status, risk classification, and molecular typing were further analyzed and compared by 21-Gene expression assay and MammaPrint plus BluePrint test.
Results: The majority of cases in both groups were invasive carcinoma (NOS), with histological grade II, HR+, Ki-67 ≥ 20%, HER2 2+, and a high risk of recurrence, although younger patients and lymph node metastases were more common in Group A. Surprisingly, all HR+ breast cancers in both groups were classified as luminal-type, HR− cases were all basal-type or unknown, and the index of HER2 in all cases was < 0.000 using the BluePrint test, which indicated that HER2 status should be negative. Furthermore, the level of HER2 mRNA expression in all cases of both groups was < 10.7, which was defined as HER2 negative by the 21-Gene expression assay. In addition, 10 patients of Group A received anti-HER2 neoadjuvant therapy; only one patient with HR- achieved Grade 5 based on the Miller-Payne system, whereas none of the patients achieved pathological complete response (pCR) based on the Residual Cancer Burden system.
Conclusion: Group A breast cancer, which has always been unquestionably diagnosed as HER2 amplification, was more likely to be HER2 negative and derived less benefit from anti-HER2 neoadjuvant chemotherapy. Group A breast cancer should be distinguished from classical HER2-positive breast cancers when assessing HER2 FISH, and a larger cohort of Group A patients should be included in further studies.
Keywords: breast cancer, HER2, FISH, 21-gene expression assay, MammaPrint plus BluePrint
Graphical Abstract:
Introduction
Human epidermal growth factor receptor 2 (HER2) gene amplification and protein expression are reported in approximately 15–20% of breast cancers and are well known associated with poor prognosis and benefit from anti-HER2 targeted therapy.1–4 Therefore, the accurate determination of HER2 status in invasive breast cancer is of utmost importance.
Currently, the status of HER2 is mainly determined by immunohistochemistry (IHC) for protein overexpression, and fluorescence in situ hybridization (FISH) for gene amplification. In clinical practice, IHC assays are typically the first method adopted. HER2 FISH is required when the IHC results are equivocal. To ensure the accuracy, reproducibility, and precision of HER2 testing, the American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) have periodically issued detailed guidelines and updated recommendations for conducting and interpreting HER2 testing in breast cancer.5–7 According to the latest version in 2018 and several recent publications, almost all breast cancers with a HER2 copy number ≥6.0 are considered positive for HER2 amplification, while those with a HER2 copy number <4.0 are considered negative by combining IHC and FISH results, regardless of the ratio of HER2/CEP17.7–12 However, the HER2/CEP17 ratio must be considered for breast cancers with an average HER2 copy number ≥4.0 and <6.0. The status of HER2 amplification is diagnosed as positive when the ratio is ≥2.0 (Group A), and negative when the ratio <2.0 (Group B) with a non HER2 3+ by IHC.7,10–13
Actually, Group B breast cancers, accounting for approximately 5–10% of breast cancers, have always been the focus in the process of HER2 ISH/FISH diagnosis.14–18 The HER2 status of Group B was considered FISH equivocal according to the 2013 ASCO/CAP guidelines, which posed a big challenge to oncologists and patients due to a perceived ambivalence regarding whether to recommend HER2-targeted therapy.6 Thus, a rigorous diagnostic approach is recommended to determine the HER2 status of Group B tumors by combining HER2 FISH and IHC results in the updated 2018 ASCO/CAP HER2 testing guidelines. Briefly, if the IHC result was 3+, the final HER2 status was positive; if the IHC result was 0, 1+, or 2+, the final HER2 status was negative.7 On the basis of this principle, almost all of the Group B cases were categorized as HER2 negative.12,13,19,20 However, Group A breast cancers with an average HER2 copy number ≥4.0 and <6.0, and an HER2/CEP17 ratio ≥2.0, reported in approximately 2.1–3.7% of breast cancer,11,12 were diagnosed as positive for HER2 amplification undoubtedly according to all versions of the ASCO/CAP guidelines.5–7 Such stark judgment and guidance for subsequent targeted therapies are controversial.
In the present study, 40 breast cancer patients with an average HER2 copy number of ≥4.0 and <6.0 were collected and classified into two groups based on the HRE2/CEP17 ratio – Group A: the ratio ≥2.0 (n=22), and Group B: ratio <2.0 (n=18). Clinicopathological features, HER2 mRNA expression, risk classification, and molecular typing of both Group A and Group B breast cancers were analyzed using a 21-Gene expression assay and the MammaPrint plus BluePrint test. Furthermore, the efficacy of anti-HER2 neoadjuvant chemotherapy (NACT) was evaluated in Group A breast cancer, which has been unquestionably diagnosed as HER2 amplification for a long time.
Materials and Methods
Patient Cohort
Nineteen cases of invasive breast cancer with an average HER2 copy number ≥4.0, <6.0, and HER2/CEP17 ratio ≥2.0 (Group A) were retrospectively screened from a total of 3119 breast cancers that underwent FISH for HER2 gene amplification at Fudan University Shanghai Cancer Center from April 2019 to March 2021. A comparable number (n=15) of breast cancer patients with an average HER2 copy number ≥4.0, <6.0, and HER2/CEP17 ratio <2.0 (Group B) were randomly selected as controls. The inclusion criteria for this study included: ① Hormone receptors (HR) were positive by IHC; ② All testing was performed in core needle biopsy samples before treatment for breast cancer patients receiving neoadjuvant therapy; ③ The proportion of invasive cancer in biopsy or excision samples should be ≥50% and the proportion of ductal carcinoma in situ (DCIS) should be <10% after tumor enrichment; ④ The nucleic acid quality should meet the needs of subsequent detection; and ⑤ Cases with incomplete clinicopathological information were excluded. Risk classification and molecular typing were performed in all cases using MammaPrint and BluePrint tests, and a 21-Gene expression assay was performed in pN0-1 (0–3 positive nodes) cases. Additionally, 3 cases with HR negativity were randomly selected from each group for molecular typing using the BluePrint test. The flowchart is shown in Figure 1. Clinicopathologic features, including patient age, menopausal status, tumor size, tumor grade, histologic type, IHC results of estrogen receptor (ER), progestogen receptor (PR), HER2, Ki-67 expression, HER2 FISH results, Miller-Payne (MP) and Residual Cancer Burden (RCB) grading, which were used to evaluate the efficacy of anti-HER2 NACT,21,22 were obtained from the medical record and pathology reports. Every patient in the study signed informed written consent. Ethical approval was obtained from the Research Ethics Committee of Fudan University Shanghai Cancer Center. All study activities were conducted in accordance with the Declaration of Helsinki.
 |
Figure 1 The flow chart of risk classification and molecular typing tests in breast cancer with an average HER2 copy number ≥4.0 and <6.0. |
Immunohistochemistry (IHC) and Fluorescence in-situ Hybridization (FISH)
All 40 patients were immunohistochemically assessed for ER, PR, HER2, and Ki-67 expression. All the antibodies used in this study were purchased from Roche Ventana. Staining was performed on 4-μm-thick tissue sections using a Ventana BenchMark Ultra Autostainer (Ventana Medical System Inc., Roche, Tuscon, AZ, USA). Positive and negative controls for ER, PR, and HER2 were included on each slide. ER, PR, and HER2 IHC statuses were evaluated according to ASCO/CAP guidelines.7,23 For Ki-67 assessment, a cut-off point of 20% was used in reference to the 2013 St. Gallen International Expert Consensus.24
The status of HER2 amplification in all 40 cases was further examined by FISH using a PathVysion HER2 DNA Probe Kit (Abbott Molecular, Abbott Park, Illinois, USA). The HER2 and CEP17 signals were manually counted by two certified molecular pathologists (QMB and HL) independently. Thirty nuclei from non-overlapping areas were counted and the corresponding HER2/CEP17 ratios were calculated. When there was a conflict between the scores, another pathologist (XYZ) reviewed the slides and obtained the final result. Results were interpreted and reported in accordance with the 2018 ASCO/CAP guideline.7
21-Gene Expression Assay by RT-PCR
A 21-gene expression assay was performed in early stage, HR+, and pN0-1 breast cancer cases. The level of HER2 mRNA expression and recurrence score (RS) were determined from formalin-fixed paraffin-embedded (FFPE) tissue as previously described.25,26 Briefly, hematoxylin and eosin (HE)-stained slides were reviewed to ensure the presence of sufficient invasive breast cancer, and RNA was extracted from eight 5-μm unstained sections. The total RNA content was measured and the absence of DNA contamination was verified. Gene-specific reverse transcription was performed, followed by standardized quantitative RT-PCR in 384-well plates using the Applied Biosystems QuantStudio™ Dx Real-Time PCR System (Foster City, CA, USA). 21-gene expression levels were quantitatively analyzed, including 16 cancer-related genes (BAG1, Bcl2, CCNB1, CD68, SCUBE2, CTSL2, EstR1, GRB7, GSTM1, HER2, Ki-67, MYBL2, PR, STK15, STMY3, and SURV) and 5 reference genes (β-actin, GAPDH, GUS, RPLPO, and TFRC). The expression of each gene was measured and normalized to that of a set of five reference genes. The reference-normalized expression measurements ranged from 0 to 15, and a 1-unit increase reflected an approximately two-fold increase in the RNA expression. A tumor was HER2-negative when it was <10.7 expression units; HER2-equivocal, ≥10.7–11.4; or HER2-positive, ≥11.5.27 RS, ranging from 0 to 100, was derived from reference-normalized expression measurements of 16 cancer-related genes. Classification of recurrence risk and prediction of chemotherapy benefit were determined based on the RS value, menopausal status, and lymph nodal stage according to the National Comprehensive Cancer Network (NCCN) guidelines version 2.2022 for invasive breast cancer and a recent publication.28,29 Briefly, for postmenopausal patients with pN0-1, the addition of chemotherapy to endocrine therapy is recommended when RS ≥ 26, only endocrine therapy when RS < 26; for premenopausal patients with pN0, consider chemotherapy followed by endocrine therapy, or alternatively, ovarian function suppression combined with either tamoxifen or an AI when RS ≥ 16, only endocrine therapy when RS ≤ 15; for premenopausal patients with pN1, consider chemotherapy followed by endocrine therapy, or alternatively, ovarian function suppression combined with either tamoxifen or an AI, regardless of the RS value.
MammaPrint and BluePrint Test
Risk classification and molecular typing were performed in all 40 breast cancer cases using MammaPrint and BluePrint tests. Briefly, after RNA extraction, library preparation, and sequencing were performed following the standard procedures of the MammaPrint/BluePrint NGS kit. The quantity of pre-capture libraries and final libraries was assessed using a Bioanalyzer DNA 1000 assay and Qubit DNA HS Assay (Thermo Fisher). The libraries from 22 cases in Group A and 18 cases in Group B breast cancer were pooled for sequencing. Targeted RNA sequencing of 70 MammaPrint and 80 BluePrint signature genes was performed on an Illumina MiSeq Dx instrument using a V3 150 cycle kit. The FASTQ files generated in the lab were sent through a secure file transfer protocol server to Genecast Biotechnology Co. Ltd, which was patterned with Agendia to offer exclusive assessment rights to MammaPrint and BluePrint testing in China for analysis and interpretation following the Agendia standard pipeline.30 MammaPrint test results are reported as a MammaPrint index (MPI) that corresponds to a High Risk (MPI ≤0.000) or Low Risk (MPI > 0.000). BluePrint test results were also reported as an index. For each sample, three indices were generated, with one index for each molecular subtype (luminal-, HER2-, and basal-type index). The subtype with the highest index among the three subtypes was the categorical subtype reported for the tumors. When the three indices are all ≤0.000, the result of subtype is “unknown”.31 By combining the MammaPrint and BluePrint test results, luminal-type tumors were further stratified into luminal A-type (BluePrint: luminal-type and MammaPrint: low-risk) and luminal B-type (BluePrint: luminal-type and MammaPrint: high-risk).
Statistical Analysis
Comparisons of clinicopathological features, HER2 mRNA expression, risk classification, and molecular typing between the two groups were performed using Pearson’s chi-square test or Fisher’s exact test. All statistical tests were two-tailed, and p-values <0.05, were considered statistically significant. All analyses were performed using SPSS (version 20.0, SPSS Inc., Chicago, IL, USA).
Results
Clinicopathological Features in Group A and Group B Breast Cancers
The clinicopathological characteristics of the 40 cases of breast cancer with an average HER2 copy number ≥4.0 and <6.0 by FISH are summarized in Table 1. Representative FISH images of the two groups are shown in Figure 2. All patients were female. The mean age of the patients in Group A was 47.5 years (range 26–72 years), which was significantly younger than that in Group B, 55.7 years (range 38–72 years). Similarly, more patients (54.5%, 12/22) in Group A breast cancer were premenopausal, while more patients (77.8%, 14/18) in Group B were postmenopausal. The tumor size ranged from 0.9 to 5 cm in diameter in Group A breast cancer, 0.7–2.0 cm in Group B breast cancer, with mean diameters of 2.9 cm and 2.1 cm, respectively.
 |
Table 1 Clinicopathological Features, HER2 mRNA Expression, Risk Classification and Molecular Typing in Breast Cancer with an Average HER2 Copy Number ≥4.0 and <6.0 |
 |
Figure 2 Representative FISH images of Group A breast cancer with HER2 ≥ 4.0 and < 6.0, and HER2/CEP17 ≥ 2.0 (A) and Group B breast cancer with HER2 ≥ 4.0 and < 6.0, and HER2/CEP17 < 2.0 (B). |
Histopathologically, all the cases were invasive carcinoma, not otherwise specified (NOS). WHO grade II was predominant in both groups A (63.6%, 14/22) and B (72.2%, 13/18), and all other cases were WHO grade III. DCIS component was observed in 54.5% (12/22) of Group A and 44.4% (8/18) of Group B breast cancer, and the DCIS proportion was all less than 10% in these cases. Compared with patients in Group B, lymph node metastases were more common in Group A (72.7%, 16/22), and the number of metastatic lymph nodes was also higher in Group A. The lymph node status of pN2 was identified in 18.2% (4/22) of the patients in Group A, but none in Group B.
Immunohistochemically, the majority of cases had Ki-67 ≥20% and HER2 2+ in both groups A and B breast cancers, and there was no statistically significant difference between the two groups. None of the HER2 IHC 3+ cases were found in either group. Thus, all cases in Group A were categorized as HER2 positive, while all cases in Group B were categorized as HER2 negative according to the 2018 ASCO/CAP guidelines. Subsequently, surrogate molecular subtypes of all cases in Group A were luminal B-HER2 positive (86.4, 19/22) and HER2 overexpression (13.6%, 3/22) based on HR status, whereas 38.9% (7/18) of luminal A, 44.4% (8/18) of luminal B-HER2 negative, and 16.7% (3/18) of triple-negative tumors were identified in Group B based on HR status and Ki-67 expression, as shown in Figure 3A.
Risk Classification for the Two Group Breast Cancers by 21-Gene Assay and MammaPrint Assay
To explore the biological differences between the two groups, risk classification was analyzed by 21-Gene expression assay and MammaPrint test in early stage, HR-positive, pN0-1 breast cancer cases. As a result, although the value of the recurrence score (RS) of Group A breast cancer (39.1, range 22.0–52.7) was significantly higher than that of Group B (32.4, range 24.3–42.3), as shown in Table 1 and Figure 3B, the absolute majority of cases in both groups, 93.3% (14/15) in Group A and 86.7% (13/15) in Group B, were identified to be a high recurrence risk based on RS value, menopausal status, and lymph node status. There was no significant difference in the recurrence risk classification between the two groups (Table 1 and Figure 3C). Similarly, 73.3% (11/15) of Group A and 66.7% (10/15) of Group B were determined to be a high recurrence risk by MammaPrint analysis as shown in Table 1 and Figure 3D The coincidence rate of the above two methods was 80% (12/15) in both Group A and Group B.
Further analysis was conducted on breast cancer cases in which the recurrence risks of the two test methods were inconsistent. As shown in Table 2, there were 3 inconsistent cases in each group. The recurrence risk of all six patients was high in the 21-Gene assay, but low in the MammaPrint test. To explore the underlying reasons for the inconsistent results, the clinicopathological characteristics of these six cases and the detailed original data from the two test methods were further analyzed. The MammaPrint index (MPI) of Case 3 in both groups fell within the classification cut-off between −0.0575 and +0.0575, suggesting that the classification accuracy was less than 90%. The other four cases were found to have relatively low PR expression (≤20%), which is an important index of high recurrence risk in the 21-Gene assay.
 |
Table 2 Analysis of Breast Cancers with Inconsistent Recurrence Risk Classification Between the Two Test Methods |
Molecular Re-Typing for the Two Group Breast Cancers by BluePrint Assay
Based on the IHC and FISH results, the surrogate molecular subtype was completely different between the two groups; thus, further molecular typing was performed using the BluePrint test. Unexpectedly, all breast cancers with HR positivity in groups A and B were classified as luminal type, and all cases with HR negativity in both groups were classified as basal type or unknown by BluePrint typing analysis. Furthermore, the HER2 index in all cases was <0.000 regardless of the HER2/CEP17 ratio. By combining the MammaPrint and BluePrint test results, 18.2% (4/22) of luminal A, 68.2% (15/22) of luminal B (HER2 negative), 4.5% (1/22) of basal, and 9.1% (2/22) of unknown were identified in Group A, while 27.8% (5/18) of luminal A, 55.6% (10/18) of luminal B (HER2 negative) and 16.7% (3/18) of basal were identified in Group B. None of the HER2-type was determined in either group, as shown in Table 1 and Figure 3E. Additionally, HER2 mRNA expression was re-evaluated in both groups using a 21-Gene assay. The average HER2 mRNA expression of the patients in Group A was approximately 9.1 (range 8.4–10.5), which was very similar to that in Group B, 9.1 (range 8.0–10.6). The HER2 mRNA expression level in all cases in both groups was <10.7, which was defined as HER2 negative as shown in Table 1 and Figure 3F.
Efficacy of Anti-HER2 NACT in Group A Breast Cancer
According to the 2018 ASCO/CAP guidelines, Group A breast cancer should be defined as HER2 positive. Thus, 10 patients of Group A breast cancer received neoadjuvant therapy including anti-HER2 targeted drugs (trastuzumab and/or pertuzumab). Pathological response to treatment was evaluated using the MP and RCB systems. The evaluation results and related clinicopathological features of the ten patients are presented in Table 3. Briefly, only one HR-negative patient achieved Grade 5 (no invasive cancer cells identifiable in primary lesion) based on the MP system. All the other nine HR-positive cases showed Grade 1–4 as partial pathological response (pPR) or non-pCR based on the MP system, while none of the patients achieved pCR based on the RCB system. Relevant clinicopathological information indicated that 8 out of the 9 breast cancers with HR positivity had a high risk of recurrence according to both the 21-Gene assay and MammaPrint test.
 |
Table 3 Assessment of Pathological Responses and Clinicopathologic Features in Group A Breast Cancers Treated with Anti-HER2 NACT |
Discussion
This study focused on invasive breast cancer cases with an average HER2 copy number of ≥4.0 and <6.0, which were subsequently classified into two groups based on the HRE2/CEP17 ratio (Group A: ratio ≥2.0, and Group B: ratio <2.0). According to the latest version of the ASCO/CAP HER2 testing guidelines, all 22 cases in Group A were determined to be HER2 positive, while all 18 cases in Group B were HER2 negative based on HER2 FISH and IHC results. Given that these two groups have the same mean range of HER2 signals/cells, the CEP17 signal is what drives the differences in classification. Thus, the clinicopathological characteristics and risk classifications of the two groups were compared. The results revealed that the majority of cases in both groups were IDC grade II, Ki-67 ≥20%, and HER2 2+ with a high risk of recurrence, although younger patients and lymph node metastases were more common in Group A. Similar phenomenon has been reported by Ballard et al.11 In their study, the HER2 status of Group A was defined as “low amplified” and “equivocal” for Group B. Similar clinicopathologic characteristics were observed in these two groups, and most of these cases were HER2 negative by IHC and more likely to be HR positive. Consistently, HER2 IHC 3+ was not observed in either group in the present study. These results suggest that direct categorization of Group A breast cancer as HER2 positive might not be reasonable.
To further evaluate the HER2 gene amplification status in Group A, BluePrint test for molecular typing was performed. BluePrint, first identified by Krijgsman et al,31 has been proven to be a robust and reliable tool to identify breast cancer molecular subtypes. In the BluePrint test, the mRNA expression of 80 genes that assess functional pathways that determine the intrinsic breast cancer molecular subtypes (luminal-type, HER2-type, and basal-type) was based on the highest index (>0.000) of the three.32 Surprisingly, all breast cancers in both Group A and Group B with HR positivity were classified as luminal-type, and all HR-negative cases were basal-type or unknown, regardless of the HER2/CEP17 ratio. Furthermore, the index of HER2 in all cases in both groups A and B was <0.000, which suggested that the HER2 status of all these cases was negative. Another unexpected finding during the BluePrint testing was that 2 out of 3 cases of Group A with HR negative were defined as “unknown”, never reported in previous publications,31–34 suggesting that Group A breast cancer might not be involved during BluePrint development design and model building, leading to the inability to discriminate these cases in clinical practice. Thus, HER2 mRNA expression was detected to further evaluate the HER2 gene status in Group A breast cancer using a 21-Gene expression assay. Our results indicated that the level of HER2 mRNA expression in all cases in both groups was <10.7, which should be defined as HER2 negative according to previous studies.26,27 Droplet digital PCR (ddPCR) and targeted next-generation sequencing (NGS) were used as alternative clinical methods to analyze HER2 copy number by Yang et al, who found that all breast cancer cases with HER2/CEP17 ratio ≥2.0 and mean HER2 signals/cell ≥6.0 showed amplification by ddPCR and major copy number gain by NGS, whereas only 20% (1/5) of cases with HER2/CEP17 ratio ≥2.0, and mean HER2 signals/cell ≥4.0 and <6.0 (equivalent to Group A cases in our study) showed copy number gain by NGS and no HER2 amplification was observed by ddPCR.35 These results provide compelling evidence that Group A breast cancer is more likely to be HER2 negative.
Regarding the response to HER2 targeted therapies for Group A breast cancer, a retrospective review of the original HERA trial results and the N9831 trial indicated that there was still a benefit from trastuzumab therapy in low-amplification breast cancers (cases with an average HER2 copy number between 4 and 9 signals/cell, or HER2/CEP17 ratio between 2.0 and 5.0), and there was no statistically significant difference compared with HER2 amplified breast cancers (cases with an average HER2 copy number >9 signals/cell, or HER2/CEP17 ratio >5.0).36,37 This is why the 2013 and 2018 ASCO/CAP guidelines recommended low amplification cases as HER2 positive “without the need for further testing”. However, breast cancers with an average HER2 copy number of ≥4.0 and <6.0 were not further analyzed as a single group in the above clinical trials. Arnould et al reported a positive correlation between the level of HER2 amplification assessed using FISH and the rate of pCR to trastuzumab-based neoadjuvant therapy. They found that the pCR rate in high-amplification (mean, >10 signals/nuclei by single-color FISH) breast cancers was significantly higher than that in low-amplification (mean, 6–10 signals/nuclei) and non-amplification (mean, <6 signals/nuclei) tumors (56% vs 22% and 6%).38 Recently, Alhamar et al also demonstrated that high HER2 amplification was significantly associated with longer OS and DFS and that these patients seemed to benefit more from HER2-targeted regimens. They further categorized group 1 (according to the 2018 ASCO/CAP guideline) into three subgroups: low amplification (HER2/CEP17 2.0–2.99, HER2 average copy number 4.0–5.9), amplification (HER2/CEP17 2.0–2.99, HER2 average copy number ≥ 6), and excessive amplification (HER2/CEP17 ≥ 3.0, HER2 average copy number ≥ 4.0). After anti-HER2 neoadjuvant therapy, the pCR rate of the excessive amplification group was higher than that of the amplification and low amplification groups (23%, 6/26 vs 20%, 2/10 and 6%, 1/18, respectively), although there was no statistically significant difference, which might be due to the limited number of cases.39 In our study, 10 patients in Group A received anti-HER2 neoadjuvant therapy. Among them, only one HR-negative case achieved Grade 5, and all the other nine HR-positive cases showed Grade 1–4 as pPR or non-pCR based on the MP system. Although the number of cases in our study was relatively small, these results suggest that Group A breast cancer might have a poor response to anti-HER2 targeted therapy.
Recently, a novel HRE2-targeted antibody drug conjugate (ADC), trastuzumab deruxtecan (T-DXd), demonstrated promising preliminary antitumor activity and resulted in significantly longer progression-free and overall survival in patients with HER2-low breast cancer, which was defined as IHC 1+ or IHC 2+ and FISH negative.40–42 Patients with breast cancer with an average HER2 copy number of ≥4.0 and <6.0, which were the focus of this study, were HER2 IHC 1+ or 2+ and more likely to be negative by FISH based on our results. Thus, we hypothesized that such patients might be classified as HER2-low and benefit from HER2-targeted ADC, but further studies are needed to confirm this hypothesis in corresponding clinical trials.
In conclusion, invasive breast cancer cases with an average HER2 copy number of ≥4.0 and <6.0 showed unique clinicopathological features, with a high risk of recurrence at high frequency regardless of the ratio of HER2/CEP17. Both Group A (ratio ≥ 2.0) and B (ratio < 2.0) breast cancers were more likely to be HER2 negative and derived less benefit from anti-HER2 neoadjuvant chemotherapy, although Group A breast cancer has always been unquestionably diagnosed as HER2 amplification. These results indicate that Group A breast cancers should be distinguished from classical HER2-positive breast cancers when assessing HER2 FISH, and a larger cohort of Group A patients should be included in further studies to clarify the biological features and efficacy of different HER2-targeted therapies, including novel HER2-targeted ADC (T-DXd).
Data Sharing Statement
The dataset used and analyzed during the current study is available from the corresponding author upon reasonable request.
Acknowledgments
This research was funded by the Science and Technology Commission of Shanghai Municipality (No.19441904900), Shanghai Science and Technology Development Fund (19MC1911000), Shanghai Municipal Key Clinical Specialty (shslczdzk01301), Innovation Program of Shanghai Science and Technology Committee (20Z11900300), and Innovation Group Project of Shanghai Municipal Health Commission Grant (2019CXJQ03).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Ballinger TJ, Sanders ME, Abramson VG. Current HER2 testing recommendations and clinical relevance as a predictor of response to targeted therapy. Clin Breast Cancer. 2015;15:171–180. doi:10.1016/j.clbc.2014.11.009
2. Ross JS, Slodkowska EA, Symmans WF, et al. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14:320–368. doi:10.1634/theoncologist.2008-0230
3. Gabos Z, Sinha R, Hanson J, et al. Prognostic significance of human epidermal growth factor receptor positivity for the development of brain metastasis after newly diagnosed breast cancer. J Clin Oncol. 2006;24:5658–5663. doi:10.1200/JCO.2006.07.0250
4. Dawood S, Broglio K, Buzdar AU, et al. Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J Clin Oncol. 2010;28:92–98. doi:10.1200/JCO.2008.19.9844
5. Wolff AC, Hammond MEH, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi:10.1200/JCO.2006.09.2775
6. Wolff AC, Hammond MEH, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi:10.1200/JCO.2013.50.9984
7. Wolff AC, Hammond MEH, Allison KH, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline focused update. J Clin Oncol. 2018;36:2105–2122. doi:10.1200/JCO.2018.77.8738
8. Bhargava R, Dabbs DJ. Interpretation of human epidermal growth factor receptor 2 (HER2) in situ hybridization assays using 2013 update of American Society of Clinical Oncology/College of American Pathologists HER2 guidelines. J Clin Oncol. 2014;32:1855. doi:10.1200/JCO.2013.53.9213
9. Wang X, Teng X, Ding W, et al. A clinicopathological study of 30 breast cancer cases with a HER2/CEP17 ratio of ≥2.0 but an average HER2 copy number of <4.0 signals per cell. Mod Pathol. 2020;33:1557–1562. doi:10.1038/s41379-020-0519-y
10. Press MF, Sauter G, Buyse M, et al. HER2 gene amplification testing by fluorescent in situ hybridization (FISH): comparison of the ASCO-College of American Pathologists guidelines with FISH scores used for enrollment in breast cancer international research group clinical trials. J Clin Oncol. 2016;34:3518–3528. doi:10.1200/JCO.2016.66.6693
11. Ballard M, Jalikis F, Krings G, et al. ‘Non-classical’ HER2 FISH results in breast cancer: a multiinstitutional study. Mod Pathol. 2017;30:227–235. doi:10.1038/modpathol.2016.175
12. Li A, Bai Q, Kong H, et al. Impact of the updated 2018 American Society of Clinical Oncology/College of American Pathologists guideline for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2020;144:1097–1107. doi:10.5858/arpa.2019-0369-OA
13. Kong H, Bai Q, Li A, et al. Characteristics of HER2-negative breast cancers with FISH-equivocal status according to 2018 ASCO/CAP guideline. Diagn Pathol. 2022;17. doi:10.1186/s13000-021-01187-z
14. Guo L, Yuan P, Zhang J, et al. Analysis of molecular subtypes for the increased HER2 equivocal cases caused by application of the updated 2013 ASCO/CAP HER2 testing guidelines in breast cancer. Breast Cancer Res Treat. 2017;166:77–84. doi:10.1007/s10549-017-4397-z
15. Hanna WM, Slodkowska E, Lu F-I, et al. Comparative analysis of human epidermal growth factor receptor 2 testing in breast Cancer according to 2007 and 2013 American Society of Clinical Oncology/College of American Pathologists guideline recommendations. J Clin Oncol. 2017;35:3039–3045. doi:10.1200/JCO.2016.70.5319
16. Bethune GC, Zanten DVV, MacIntosh RF, et al. Impact of the 2013 American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 (HER2) testing of invasive breast carcinoma: a focus on tumours assessed as “equivocal” for HER2 gene amplification by fluorescence in-situ hybridization. Histopathology. 2015;67:880–887. doi:10.1111/his.12723
17. Liu Y, Wu S, Shi X, et al. HER2 double-equivocal breast cancer in Chinese patients: a high concordance of HER2 status between different blocks from the same tumor. Breast Cancer Res Treat. 2019;178:275–281. doi:10.1007/s10549-019-05387-6
18. Xu Y, Bai QM, Yang F, et al. Impact of 2013 American Society of Clinical Oncology/College of American Pathologist guidelines on borderline immunostaining results for HER2: a retrospective study on HER2 FISH results in 1780 cases of invasive breast cancers. Zhonghua Bing Li Xue Za Zhi. 2016;45:545–549. doi:10.3760/cma.j.issn.0529-5807.2016.08.010
19. Press MF, Villalobos I, Santiago A, et al. Assessing the new American Society of Clinical Oncology/College of American Pathologists guidelines for HER2 testing by fluorescence in situ hybridization: experience of an academic consultation practice. Arch Pathol Lab Med. 2016;140:1250–1258. doi:10.5858/arpa.2016-0009-OA
20. Shah MV, Wiktor AE, Meyer RG, et al. Change in pattern of HER2 fluorescent in situ hybridization (FISH) results in breast cancers submitted for FISH testing: experience of a reference laboratory using US Food and Drug Administration criteria and American Society of Clinical Oncology and College of American Pathologists guidelines. J Clin Oncol. 2016;34:3502–3510. doi:10.1200/JCO.2015.61.8983
21. Ogston KN, Miller ID, Payne S, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–327. doi:10.1016/S0960-9776(03)00106-1
22. Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25:4414–4422. doi:10.1200/JCO.2007.10.6823
23. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi:10.1200/JCO.2009.25.6529
24. Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi:10.1093/annonc/mdt303
25. Cronin M, Sangli C, Liu ML, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53:1084–1091. doi:10.1373/clinchem.2006.076497
26. Qi P, Yang Y, Bai QM, et al. Concordance of the 21-gene assay between core needle biopsy and resection specimens in early breast cancer patients. Breast Cancer Res Treat. 2021;186:327–342. doi:10.1007/s10549-020-06075-6
27. Park MM, Ebel JJ, Zhao W, et al. ER and PR immunohistochemistry and HER2 FISH versus oncotype DX: implications for breast cancer treatment. Breast J. 2014;20:37–45. doi:10.1111/tbj.12223
28. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–121. doi:10.1056/NEJMoa1804710
29. Kalinsky K, Barlow WE, Gralow JR, et al. 21-gene assay to inform chemotherapy benefit in node-positive breast cancer. N Engl J Med. 2021;385:2336–2347. doi:10.1056/NEJMoa2108873
30. Mittempergher L, Delahaye LJ, Witteveen A, et al. MammaPrint and BluePrint molecular diagnostics using targeted RNA next-generation sequencing technology. J Mol Diagn. 2019;21:808–823. doi:10.1016/j.jmoldx.2019.04.007
31. Krijgsman O, Roepman P, Zwart W, et al. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat. 2012;133:37–47. doi:10.1007/s10549-011-1683-z
32. Mittempergher L, Delahaye LJ, Witteveen AT, et al. Performance characteristics of the BluePrint® breast cancer diagnostic test. Transl Oncol. 2020;13:100756. doi:10.1016/j.tranon.2020.100756
33. Whitworth P, Stork-Sloots L, de Snoo FA, et al. Chemosensitivity predicted by BluePrint 80-gene functional subtype and MammaPrint in the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST). Ann Surg Oncol. 2014;21:3261–3267. doi:10.1245/s10434-014-3908-y
34. Viale G, de Snoo FA, Slaets L, et al. Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial. Breast Cancer Res Treat. 2018;167:123–131. doi:10.1007/s10549-017-4509-9
35. Yang SR, Bouhlal Y, De La Vega FM, et al. Integrated genomic characterization of ERBB2/HER2 alterations in invasive breast carcinoma: a focus on unusual FISH groups. Mod Pathol. 2020;33:1546–1556. doi:10.1038/s41379-020-0504-5
36. Dowsett M, Procter M, McCaskill-Stevens W, et al. Disease-free survival according to degree of HER2 amplification for patients treated with adjuvant chemotherapy with or without 1 year of trastuzumab: the HERA Trial. J Clin Oncol. 2009;27:2962–2969. doi:10.1200/JCO.2008.19.7939
37. Perez EA, Reinholz MM, Hillman DW, et al. HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. J Clin Oncol. 2010;28:4307–4315. doi:10.1200/JCO.2009.26.2154
38. Arnould L, Arveux P, Couturier J, et al. Pathologic complete response to trastuzumab-based neoadjuvant therapy is related to the level of HER-2 amplification. Clin Cancer Res. 2007;13:6404–6409. doi:10.1158/1078-0432.CCR-06-3022
39. Alhamar M, Alkamachi B, Mehrotra H, et al. Clinical significance of quantitative categorization of HER2 fluorescent in situ hybridization results in invasive breast cancer patients treated with HER2-targeted agents. Mod Pathol. 2021;34:720–734. doi:10.1038/s41379-020-00728-z
40. Modi S, Jacot W, Yamashita T, et al. Trastuzumab Deruxtecan in previously treated HER2-low advanced breast cancer. N Engl J Med. 2022;387:9–20. doi:10.1056/NEJMoa2203690
41. Denkert C, Seither F, Schneeweiss A, et al. Clinical and molecular characteristics of HER2-low-positive breast cancer: pooled analysis of individual patient data from four prospective, neoadjuvant clinical trials. Lancet Oncol. 2021;22:1151–1161. doi:10.1016/S1470-2045(21)00301-6
42. Modi S, Park H, Murthy RK, et al. Antitumor activity and safety of Trastuzumab Deruxtecan in patients with HER2-low-expressing advanced breast cancer: results from a phase Ib study. J Clin Oncol. 2020;38:1887–1896. doi:10.1200/JCO.19.02318
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

