Back to Journals » Infection and Drug Resistance » Volume 15
Introduction of Probiotic-Based Sanitation in the Emergency Ward of a Children’s Hospital During the COVID-19 Pandemic
Authors Soffritti I, D'Accolti M, Cason C, Lanzoni L, Bisi M, Volta A, Campisciano G, Mazzacane S, Bini F, Mazziga E, Toscani P, Caselli E , Comar M
Received 12 January 2022
Accepted for publication 16 March 2022
Published 30 March 2022 Volume 2022:15 Pages 1399—1410
DOI https://doi.org/10.2147/IDR.S356740
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Irene Soffritti,1,2 Maria D’Accolti,1,2 Carolina Cason,3 Luca Lanzoni,2 Matteo Bisi,2 Antonella Volta,2 Giuseppina Campisciano,3 Sante Mazzacane,2 Francesca Bini,1,2 Eleonora Mazziga,1 Paola Toscani,4 Elisabetta Caselli,1,2,* Manola Comar3,5,*
1Section of Microbiology, Department of Chemical, Pharmaceutical and Agricultural Sciences, and LTTA, University of Ferrara, Ferrara, 44121, Italy; 2CIAS Research Center, University of Ferrara, Ferrara, 44122, Italy; 3Department of Advanced Translational Microbiology, Institute for Maternal and Child Health-IRCCS “Burlo Garofolo”, Trieste, 34137, Italy; 4Institute for Maternal and Child Health-IRCCS “Burlo Garofolo”, Trieste, 34137, Italy; 5Department of Medical Sciences, University of Trieste, Trieste, 34149, Italy
*These authors contributed equally to this work
Correspondence: Elisabetta Caselli, Section of Microbiology, Department of Chemical, Pharmaceutical and Agricultural Sciences, University of Ferrara, via Luigi Borsari 46, Ferrara, 44121, Italy, Tel +39 0532 455387, Fax +39 0532 974470, Email [email protected]
Background: Antimicrobial resistance (AMR) represents a major threat to public health, especially in the hospital environment, and the massive use of disinfectants to prevent COVID-19 transmission might intensify this risk, possibly leading to future AMR pandemics. However, the control of microbial contamination is crucial in hospitals, since hospital microbiomes can cause healthcare-associated infections (HAIs), which are particularly frequent and severe in pediatric wards due to children having high susceptibility.
Aim: We have previously reported that probiotic-based sanitation (PCHS) could stably decrease pathogens and their AMR in the hospital environment, reduce associated HAIs in adult hospitals, and inactivate enveloped viruses. Here, we aimed to test the effect of PCHS in the emergency room (ER) of a children’s hospital during the COVID-19 pandemic.
Methods: Conventional chemical disinfection was replaced by PCHS for 2 months during routine ER sanitation; the level of environmental bioburden was characterized before and at 2, 4, and 9 weeks after the introduction of PCHS. Microbial contamination was monitored simultaneously by conventional culture-based CFU count and molecular assays, including 16S rRNA NGS for bacteriome characterization and microarrays for the assessment of the resistome of the contaminating population. The presence of SARS-CoV-2 was also monitored by PCR.
Results and conclusions: PCHS usage was associated with a stable 80% decrease in surface pathogens compared to levels detected for chemical disinfection (P < 0.01), accompanied by an up to 2 log decrease in resistance genes (Pc < 0.01). The effects were reversed when reintroducing chemical disinfection, which counteracted the action of the PCHS. SARS-CoV-2 was not detectable in both the pre-PCHS and PCHS periods. As the control of microbial contamination is a major issue, especially during pandemic emergencies, collected data suggest that PCHS may be successfully used to control virus spread without simultaneous worsening of the AMR concern.
Keywords: drug resistance, cross infection, biological monitoring, COVID-19, probiotics
Introduction
The ongoing COVID-19 pandemic has profoundly affected our environmental hygiene habits, due to the ability of SARS-CoV-2 to persist on surfaces for a long period of time.1–3 Although fomite transmission is difficult to prove definitively, sanitization measures based on the massive use of chemical disinfectants have been mandatorily introduced worldwide, in both healthcare and non-healthcare settings.4–6 Indeed, disinfectants’ action is temporary,7,8 allowing the treated surface to be rapidly recontaminated and potentially serve as a source of infection. Furthermore, the widespread use of disinfectants may be itself a threat to human health, negatively affecting water and earth pollution,9,10 and potentially inducing further increases in the antimicrobial resistance (AMR) of pathogens known to complicate COVID‐19 clinical care.11–15 Considering the current spread of AMR, responsible for the death of over 37,000 people/year in Europe alone,16 the risk of future AMR pandemics has been recently highlighted by WHO.17 AMR is particularly frequent in healthcare settings, due to the selective pressure exerted by the continuous use of disinfectants and antibiotics, where it is closely associated with the severity of healthcare-associated infections (HAIs),18,19 affecting up to 15% of all hospitalized inpatients,20 with mortality rates reaching up to 30% in Italy.21 HAI incidence is particularly high in pediatric wards, due to the high susceptibility of children to infections.22 Premature babies are particularly exposed to the risk of contracting uncommon and rare HAIs,23 and we recently reported colonization of preterm newborns by environmental Intensive Care Unit (ICU)-resistant microbes,24 supporting the significant potential contribution of environmental hospital microbes in children with HAIs.
The hospital microbiome represents a reservoir of pathogens listed by WHO in the ESKAPE and Dirty Dozen groups, including microbes which can survive on surfaces for a long period of time and be transmitted to hospital inpatients;25–27 these are dangerous primarily because of their drug-resistance.6 Hospital cleaning plays a crucial role in reducing bioburden and HAI risk. Due to the limitations of chemical disinfection, an efficient, low‐impact, and possibly low‐cost method would be needed that is able to ensure long-lasting sanitation of treated surfaces as well as being able to overcome undesirable side effects. Toward this direction, we studied a probiotic-based method, showing that it can induce a stable 80% decrease in environmental pathogens compared to disinfectants,7,28 an up to 99.9% decrease in resistance genes,29 and a concomitant >50% reduction in HAI incidence,30 similar to results emerging from studies performed in the Internal Medicine wards of several public hospitals in Italy and Belgium. Such a system consists of an eco-friendly detergent containing spores of probiotic Bacilli that can replace pathogens on treated surfaces (Probiotic Cleaning Hygiene System, PCHS) through its competitive exclusion mechanism and the production of antimicrobial compounds.31 PCHS is characterized by ease of use and low cost, and has recently been proven to inactivate enveloped viruses, including SARS-CoV-2, keeping the treated surfaces protected for up to 24 hours.8
On the other hand, monitoring the hospital environment through rapid and precise methods can provide essential information to efficiently control pathogenic HAI-associated bioburden in pediatric hospitals.32 Thus, we characterized the microbial contamination of the emergency ward of an Italian children’s hospital during the COVID-19 pandemic, and assessed the effectiveness of PCHS sanitation compared to chemical sanitation.
Materials and Methods
Study Design and Environmental Sampling
The study was conducted in the emergency rooms (ERs) of the Maternal and Child Health Institute “IRCCS Burlo Garofolo” (Trieste, Italy), after obtaining approval from the Institutional Review Board of IRCCS Burlo Garofolo (RC18/17). Conventional chemical disinfection was replaced by PCHS for 2 months (Probiotic Cleaning Hygiene System, Copma scrl, Ferrara, Italy). The PCHS consisted of a patented eco-friendly detergent with the addition of spores of three selected probiotic bacteria of the Bacillus genus (B. subtilis, B. pumilus, and B. megaterium). PCHS sanitation was applied twice a day—in the early morning and afternoon—as also routinely applied for chemical sanitation. All the other Infections Prevention and Control (IPC) procedures were left unaltered, as carried out in the previous studies.30 Emergency disinfection with 0.5% sodium hypochlorite (NaClO) was permitted in cases of confirmed SARS-CoV-2 positivity of subjects admitted to the ER. Five sampling campaigns were performed before and after PCHS introduction: two campaigns in the pre-PCHS period (T01, T02), with a 10-day interval, and three campaigns in the PCHS period (T1, T2, T3), at 2, 5 and 9 weeks after PCHS introduction. Floor, bed footboard, and sink surfaces were sampled in duplicate 7 hours after cleaning,7,28,30 in the following areas: triage room, corridor, refectory room, COVID-19 suspected patients’ room, and three hospitalization rooms. The surfaces were simultaneously sampled by two different methods (contact plates or sterile swabs), according to subsequent microbiological or molecular analyses (Figure 1).
 |
Figure 1 Conceptual framework of the used methodology. |
Microbiological Analyses
Surface microbial contamination was analyzed by Replicate Organism Detection and Counting contact plates (RODAC, Merck Millipore, Milan, Italy), measuring: total bacteria (TSA medium), Staphylococcus spp. (Baird Parker medium), Enterobacteriaceae spp. (MacConkey medium), Acinetobacter spp. (Herella medium), Pseudomonas spp. (Cetrimide medium), Clostridium difficile (Clostridium difficile medium), Enterococcus spp. (BEA medium) and mycetes (Sabouraud medium). Briefly, duplicate samples were collected from surfaces and incubated anaerobically (Clostridium medium) or aerobically (all the remaining media) for 24–48 h (bacteria), or at 25 °C for 72 h (mycetes).24,32 At the end of incubation, colony-forming units (CFU) were counted, and S. aureus identification was performed on Baird–Parker medium and confirmed by API Staph (bioMerieux, Inc, Durham, NC, USA), as previously described.7
Molecular Analyses
The whole bacteriome, the microbial resistome, and the presence of SARS-CoV-2 genome were assessed by molecular analyses. For this purpose, samples were collected in duplicate by sterile rayon swabs (Copan, Brescia, Italy), premoistened in sterile saline and rubbed on a 10×10 cm (100 cm2) area. Swabs were then placed in 0.3 mL sterile saline and frozen at −80°C until use, as previously described.7,24,28,32 Total nucleic acids were extracted using the Maxwell HT Viral TNA automatic extractor kit (Promega, Madison, WI, USA), following the manufacturer’s instructions.
The bacteriome was characterized by Next Generation Sequencing (NGS), by sequencing the V3 region of the 16S rRNA gene and including negative controls with no DNA template, as previously described.22,30–33 Briefly, a quantitative real time PCR (qPCR) was first performed to amplify all the bacterial species by using degenerated primers; then, a nested PCR targeting the V3 region of the 16S rRNA gene was performed, as already described.24 The obtained sequences were processed using Quantitative Insights Into Microbial Ecology (QIIME 2 2020.2).24 Taxonomy assignment at the genus level was performed against the reference taxonomy database SILVA V.132,33 with a similarity threshold of 97%.
The microbial resistome was analyzed by the Microbial DNA qPCR Array for Antibiotic Resistance Genes (Qiagen, Hilden, Germany), facilitating the detection and quantification of 84 AMR genes, as previously described.7,28
The presence of the SARS-CoV-2 RNA genome was assessed by the NeoPlexTM COVID-19 detection kit (GeneMatrix, Seongnam, Gyeonggi-do, South Korea), targeting the viral N and RdRp genes, following the manufacturer’s instructions.
Statistical Analyses
Statistical analyses were performed with QIIME 2 (2020.2) and GraphPad software. Analyses of Similarities (ANOSIM) and Kruskal–Wallis tests were performed to compare community composition in the groups (T0, T1, T2, T3), assuming the P value as statistically significant if ≤ 0.05. For CFU count, parametric Student’s t-test and ANOVA test were used for group comparison, considering a P value ≤ 0.05 as significant. For resistome data, which were obtained via qPCR microarray simultaneously analyzing 84 resistance genes, the Bonferroni correction was applied for multiple comparisons, assuming a corrected Pc value ≤ 0.05 as statistically significant.
Results
Characterization of the ER’s Environmental Contamination Before and After PCHS Introduction
Monitored ER areas, before and after PCHS introduction, included the triage room, corridor, refectory room, COVID-19 suspected patients’ room, and three hospitalization rooms; 152 samples in total were collected from floor, sink and bed footboard surfaces, representing critical points in bioburden monitoring.7,30
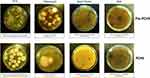
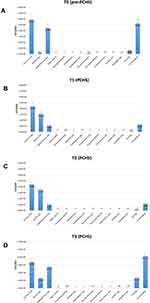
The search for common HAI-associated microorganisms was performed using RODAC plate culture, and included Staphylococcus spp., Enterobacteriaceae spp., Acinetobacter spp., Pseudomonas spp., Clostridium, Enterococcus spp., and mycetes. Representative pictures of incubated RODAC plates before and after PCHS introduction are depicted in Figure 2. The analysis of results showed evident pathogen contamination at the basal level (Figure 3A). At T0, the total amount of pathogens corresponded to 26,315 CFU/m2 (median value, 95% CI 19,155–52,334), representing 89.1% of the total microbial contamination (29,523 CFU/m2 median value, 95% CI 21,533–56,924). The Staphylococcus genus was the most prevalent, representing 85.1% of the total pathogens (22,390 CFU/m2, 95% CI 16,045–49,554). Mycetes were also detectable, representing approximately 12.0% of the total pathogens (3170 CFU/m2, 95% CI 1189–9124). Bacillus spp. were instead scarcely present at T0, with 991 CFU/m2 (95% CI 0–1448), corresponding to 3.3% of the total microbial population.
After 2 weeks of PCHS usage (T1; Figure 3B), Bacillus spp. were increased to 15,418 CFU/m2 (95% CI 12,498–21,501), representing 69.9% of the total microbial population measured. Meanwhile, the sum of total pathogens dropped to 6365 CFU/m2 (95% CI 4555–10,201), corresponding to 28.9% of the total microbes at T1, and to 24.2% of the original amount at T0. Staphylococcus spp. diminished to 5598 CFU/m2 (95% CI 3478–8223), still representing the most prevalent pathogens (87.9% of the T1 total pathogens), but representing only 25.4% of the T1 total microbial population. Mycetes also decreased to 533 CFU/m2 (95% CI 0–1109), representing 2.4% of the T1 population and 16.8% of the original T0 value, respectively. Different surfaces demonstrated diverse levels of contamination, with the highest level identified on the floor surface, followed by sink and bed footboard surfaces (Figure 4A). The total pathogens further decreased at T2 (5 PCHS weeks; Figure 3C), but not at T3 (9 PCHS weeks; Figure 3D), when their amount was comparable to pre-PCHS (T0). To understand the reason for pathogen regrowth, we first assessed the amount of PCHS Bacillus on treated surfaces (Figure 4B), showing an evident decrease at T3 compared to T1 and T2. To elucidate the possible causes of Bacillus decrease, we assessed the frequency of usage of emergency disinfection, which was allowed in cases of confirmed SARS-CoV-2 positivity in ER patients. The results (Figure 5) indicated that starting from one week after T2, chemical disinfection with 0.5% NaClO was used almost daily, due to concerns related to an increased number of SARS-CoV-2-positive children, likely inactivating the PCHS Bacillus and preventing the action of the PCHS.
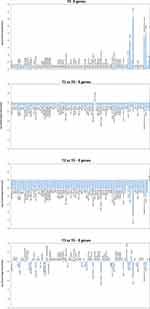
As the results of culture-based methods are limited to the searched species, the whole ER bacteriome was simultaneously characterized by NGS. The results (Figure 6) showed that Staphylococcus spp. relative abundance significantly diminished in the PCHS period (T1: 9%; T2: 9%; T3: 8%) compared to the pre-PCHS period (T0, 19%; P < 0.001). Streptococcus spp. also diminished significantly (T0: 8%; T1: 6%; T2: 3%; T3: 2%; P < 0.001), together with the less abundant pathogens Escherichia spp. and Shigella spp., showing a 2% relative abundance at T0 (P < 0.01). Acinetobacter spp. (T0: 12%; T1: 22%; T2: 12%; T3: 14%) and Pseudomonas spp. (T0: 20%; T1: 21%; T2: 30%; T3: 12%) showed no statistically significant variations over time, and Lactobacillus spp. showed slightly insignificant increasing values of relative abundance over time (T0: 1%; T1: 3%; T2: 2%; T3: 5%).
Characterization of the Environmental Resistome
The resistome microarray analysis showed that the ER microbiome harbored several AMR genes at T0 (Figure 7). In particular, the msrA, ermC, ermB, and mecA genes, providing resistance against macrolides, erythromycin and methicillin, were detected at high levels. The aadA1, aphA6, GES, IMI, Per-2, VIM-1, MIR, OXA-23, OXA-51, QnrA, QnrB-1, QnrD, and tetA genes, conferring resistance to streptomycin/spectinomycin, aminoglycoside, β-lactams, fluoroquinolones and tetracyclin, respectively, were also detected. Following PCHS introduction, a significant decrease in resistance genes was observed at T1 (Pc < 0.01) and T2 (Pc < 0.001), whereas no significant variations were observed at T3 compared to the original values at T0.
Analysis of SARS-CoV-2 Presence in ER Areas
The presence of the SARS-CoV-2 genome in the ER areas was assessed by qRT-PCR, showing no positive environmental samples during either the pre-PCHS or the PCHS period.
Discussion
The excessive use of disinfectants introduced in both healthcare and non-healthcare settings to control COVID-19 spread might lead to worsening of the AMR situation, which is already a major threat to human health,17 and is associated with the severity of HAIs, which can be particularly frequent in pediatric wards.22,23 However, the control of microbial contamination, including SARS-CoV-2 presence, is a key point to address for the prevention of colonization and infection of patients, especially in critical care pediatric wards, where very young patients are particularly susceptible.24,30 Consistent with this, we aimed to assess the potential of an innovative probiotic-based sanitation system (Probiotic Cleaning Hygiene System, PCHS; Copma scrl, Ferrara, Italy) for controlling levels of bacterial, fungal, and viral bioburden in a children’s hospital, without intensifying the threat of AMR. The action of the PCHS has previously been tested in the internal medicine wards of adult hospitals, demonstrating its ability to stably decrease the levels of pathogens, AMR, and HAI incidence,29,30,34 and in vitro, showing its capacity to inactivate enveloped viruses.8 Thus, in this study, the effects of PCHS usage were investigated for the first time in a pediatric hospital, in an emergency room setting, during the COVID-19 pandemic.
Briefly, PCHS replaced the conventional chemical sanitation protocol for a 2-month period and the level of environmental bioburden was monitored every two weeks before and after PCHS introduction, to assess any variation in load and composition, including the presence of SARS-CoV-2.
The results showed substantial pathogen contamination in the pre-PCHS period (> 26,000 CFU/m2), with a prevalent contamination by Staphylococcus spp. (> 22,000 CFU/m2) and the simultaneous presence of several other HAI-associated pathogens, including fungi (> 3000 CFU/m2), Pseudomonas spp., Clostridium difficile, and Enterococcus spp.
In parallel, the resistome analysis of the contaminant population in the pre-PCHS period provided evidence for the high prevalence of methicillin-resistant Staphylococci, and the presence of R genes, conferring resistance against several antibiotics, including macrolides, erythromycin, methicillin, streptomycin/spectinomycin, aminoglycoside, β-lactams, fluoroquinolones and tetracyclin.
The replacement of disinfectants by PCHS induced an evident microbiome remodulation in the treated areas within 2 weeks, with a stable > 80% abatement of the whole population of HAI-associated pathogens, thus confirming previously obtained results in adult healthcare settings.28,30 Furthermore, the action of PCHS did not select any drug-resistant strain; instead, it induced an up to 2 Log decrease in all the resistance genes detected in the pre-PCHS period.
Of note, the effect of PCHS was, however, reversed by the simultaneous daily use of bactericidal/sporicidal chemical disinfections, likely by inactivating the PCHS Bacillus probiotic component, confirming that the effect of PCHS is primarily based on the action carried out by the germinated Bacillus species, which are able to produce antimicrobial compounds and effectively compete with pathogens.
Lastly, no SARS-CoV-2-positive environmental samples were detected in the pre-PCHS or PCHS periods of the study, despite the documented presence of SARS-CoV-2-positive subjects, confirming the low risk of virus transmission via surfaces35 and corroborating the proven in vitro capability of PCHS to inactivate enveloped viruses.8
Limitations of this study include the monocentric design of the research and the limited time period of PCHS application, since increasing the time of analysis and the number of settings would allow generalization of the results. In addition, a prolonged time of analysis would also permit the assessment of the impact of PCHS implementation on HAI incidence, of which there is still a lack of data for pediatric hospitals. In light of the high susceptibility of hospitalized children to infections and of the high mortality rate of such infections, especially in low-income countries, this could provide important comparative results useful for evaluating the potential effects of probiotic-based versus chemical sanitation procedures.
Conclusion
The present study provided, for the first time, the characterization of bioburden level in the emergency room of an Italian children’s hospital using the chemical disinfection required during the COVID-19 pandemic, demonstrating the persistent presence of several drug-resistant HAI-associated pathogens. The introduction of PCHS sanitation stably decreased the amount of pathogens present and AMR, while keeping the environment free of SARS-CoV-2. Thus, these collected data may open the way to considering such a method as a potential effective and eco-friendly alternative to conventional chemical sanitation.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chin AWH, Poon LLM. Stability of SARS-CoV-2 in different environmental conditions - authors’ reply. Lancet Microbe. 2020;1(4):e146. doi:10.1016/S2666-5247(20)30095-1
2. Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104(3):246–251. doi:10.1016/j.jhin.2020.01.022
3. Marques M, Domingo JL. Contamination of inert surfaces by SARS-CoV-2: persistence, stability and infectivity. A review. Environ Res. 2021;193:110559. doi:10.1016/j.envres.2020.110559
4. CDC CfDCaP. SARS-CoV-2 and surface (fomite) transmission for indoor community environments; 2021. Available from: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/surface-transmission.html#ref19.
5. ISS ISdS. Interim recommendations on cleaning and disinfection of non-healthcare settings during COVID-19 health emergency: surfaces, indoor environments and clothing. In: ISS COVID-19 Working Group on Biocides. ISS; Rapporto ISS COVID-19 n. 25/2020 (in Italian). 2020.
6. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics; 2017. Available from: http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
7. Vandini A, Temmerman R, Frabetti A, et al. Hard surface biocontrol in hospitals using microbial-based cleaning products. PLoS One. 2014;9(9):e108598. doi:10.1371/journal.pone.0108598
8. D’Accolti M, Soffritti I, Bonfante F, Ricciardi W, Mazzacane S, Caselli E. Potential of an eco-sustainable probiotic-cleaning formulation in reducing infectivity of enveloped viruses. Viruses. 2021;13(11):2227. doi:10.3390/v13112227
9. Nabi G, Wang Y, Hao Y, Khan S, Wu Y, Li D. Massive use of disinfectants against COVID-19 poses potential risks to urban wildlife. Environ Res. 2020;188:109916. doi:10.1016/j.envres.2020.109916
10. Zhang H, Tang W, Chen Y, Yin W, Sills J. Disinfection threatens aquatic ecosystems. Science. 2020;368(6487):146–147. doi:10.1126/science.abb8905
11. Knight GM, Glover RE, McQuaid CF, et al. Antimicrobial resistance and COVID-19: interSections and implications. Elife. 2021;10. doi:10.7554/eLife.64139
12. Kampf G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics. 2018;7(4). doi:10.3390/antibiotics7040110
13. Wand ME, Bock LJ, Bonney LC, Sutton JM. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother. 2017;61(1). doi:10.1128/AAC.01162-16
14. Zeshan B, Karobari MI, Afzal N, et al. The usage of antibiotics by COVID-19 patients with comorbidities: the risk of increased antimicrobial resistance. Antibiotics. 2021;11(1). doi:10.3390/antibiotics11010035
15. Karobari MI, Khijmatgar S, Bhandary R, et al. A multicultural demographic study to analyze antibiotic prescription practices and the need for continuing education in dentistry. Biomed Res Int. 2021;2021:5599724. doi:10.1155/2021/5599724
16. ECDC. Surveillance of antimicrobial resistance in Europe – 2017; 2017. Available from: www.ecdc.europa.eu.
17. Getahun H, Smith I, Trivedi K, Paulin S, Balkhy HH. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ. 2020;98:442–442A. doi:10.2471/BLT.20.268573
18. Caini S, Hajdu A, Kurcz A, Böröcz K. Hospital-acquired infections due to multidrug-resistant organisms in Hungary, 2005–2010. Euro Surveill. 2013;18:20352.
19. Cornejo-Juarez P, Vilar-Compte D, Perez-Jimenez C, Namendys-Silva SA, Sandoval-Hernandez S, Volkow-Fernandez P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int J Infect Dis. 2015;31:31–34. doi:10.1016/j.ijid.2014.12.022
20. Allegranzi B, Bagheri Nejad S, Combescure C, et al. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. doi:10.1016/S0140-6736(10)61458-4
21. Messineo A, Marsella LT. Biological hazards and healthcare-associated infections in Italian healthcare facilities: some considerations on inspections and accountability. Ann Ig. 2015;27(6):799–807. doi:10.7416/ai.2015.2073
22. Lake JG, Weiner LM, Milstone AM, Saiman L, Magill SS, See I. Pathogen distribution and antimicrobial resistance among pediatric healthcare-associated infections reported to the national healthcare safety network, 2011–2014. Infect Cont Hosp Epidemiol. 2018;39(1):1–11. doi:10.1017/ice.2017.236
23. Wagh A, Sinha A. Prevention of healthcare-associated infections in paediatric intensive care unit. Childs Nerv Syst. 2018;34(10):1865–1870. doi:10.1007/s00381-018-3909-4
24. Cason C, D’Accolti M, Campisciano G, et al. Microbial contamination in hospital environment has the potential to colonize preterm newborns’ nasal cavities. Pathogens. 2021;10(5):615. doi:10.3390/pathogens10050615
25. Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi:10.1186/1471-2334-6-130
26. Otter JA, Yezli S, Salkeld JAG, French GL. Evidence that contaminated surfaces contribute to the transmission of hospital pathogens and an overview of strategies to address contaminated surfaces in hospital settings. Am J Infect Control. 2013;41(5):S6–S11. doi:10.1016/j.ajic.2012.12.004
27. Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RLP, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45(8):992–998. doi:10.1086/521854
28. Caselli E, D’Accolti M, Vandini A, et al. Impact of a probiotic-based cleaning intervention on the microbiota ecosystem of the hospital surfaces: focus on the resistome remodulation. PLoS One. 2016;11(2):e0148857. doi:10.1371/journal.pone.0148857
29. Caselli E, Arnoldo L, Rognoni C, et al. Impact of a probiotic-based hospital sanitation on antimicrobial resistance and HAI-associated antimicrobial consumption and costs: a multicenter study. Infect Drug Resist. 2019;12:501–510. doi:10.2147/IDR.S194670
30. Caselli E, Brusaferro S, Coccagna M, et al. Reducing healthcare-associated infections incidence by a probiotic-based sanitation system: a multicentre, prospective, intervention study. PLoS One. 2018;13(7):e0199616. doi:10.1371/journal.pone.0199616
31. D’Accolti M, Soffritti I, Mazzacane S, Caselli E. Fighting AMR in the healthcare environment: microbiome-based sanitation approaches and monitoring tools. Int J Mol Sci. 2019;20(7):1535. doi:10.3390/ijms20071535
32. Comar M, D’Accolti M, Cason C, et al. Introduction of NGS in environmental surveillance for healthcare-associated infection control. Microorganisms. 2019;7(12):708. doi:10.3390/microorganisms7120708
33. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–596. doi:10.1093/nar/gks1219
34. Caselli E, D’Accolti M, Soffritti I, et al. An innovative strategy for the effective reduction of MDR pathogens from the nosocomial environment. Adv Exp Med Biol. 2019;1214:79–91.
35. D’Accolti M, Soffritti I, Passaro A, et al. SARS-CoV-2 RNA contamination on surfaces of a COVID-19 ward in a hospital of Northern Italy: what risk of transmission? Eur Rev Med Pharmacol Sci. 2020;24(17):9202–9207. doi:10.26355/eurrev_202009_22872
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.