Back to Journals » Cancer Management and Research » Volume 11
Integrin α5/ITGA5 Promotes The Proliferation, Migration, Invasion And Progression Of Oral Squamous Carcinoma By Epithelial–Mesenchymal Transition
Received 14 July 2019
Accepted for publication 5 October 2019
Published 13 November 2019 Volume 2019:11 Pages 9609—9620
DOI https://doi.org/10.2147/CMAR.S223201
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Xueqiong Zhu
Yun Deng,1 Quan Wan,2 Wangxiang Yan1
1Department of Stomatology, First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, People’s Republic of China; 2Department of Oral and Maxillofacial Surgery, Sun Yat-Sen Memorial Hospital, Guangzhou, People’s Republic of China
Correspondence: Wangxiang Yan
Department of Stomatology, First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong 510000, People’s Republic of China
Email [email protected]
Background: Integrin signalling is involved in cell migration, invasion, proliferation and motility. Integrin α5/ITGA5 is a subunit of Integrin and contributes to the activation of Integrin signalling. The potential role of Integrin α5/ITGA5 in oral squamous cancer remains unknown. The aim of this study was to uncover the effect and mechanism of Integrin α5/ITGA5 in the progression of oral squamous carcinoma.
Method: TCGA database scanning, qRT-PCR, immunohistochemistry and Western blotting assays were used to detect the expression of Integrin α5/ITGA5 in tissues and cell lines. We established stable Integrin α5/ITGA5 overexpressing and Integrin α5/ITGA5 knockdown cell lines. We investigated the biological function and the underlying mechanism of Integrin α5/ITGA5 through a series of experiments.
Results: Integrin α5/ITGA5 was upregulated in cancer tissue, and its levels negatively correlated with the overall survival (OS) of patients. Integrin α5/ITGA5 promoted proliferation, migration and invasion in an oral squamous carcinoma cell line by EMT (epithelial–mesenchymal transition).
Conclusion: Integrin α5/ITGA5 promotes the proliferation, migration and invasion of oral squamous carcinoma.
Keywords: oral squamous carcinoma, Integrin α5/ITGA5, EMT
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of cancer worldwide.1 OSCC is associated with poor prognosis and high morbidity when diagnosed at advanced stages, and it is considered to be a very immunosuppressive cancer.2 Despite advances in radiotherapy and surgical therapy, patients with late-stage OSCC still suffer from metastasis and recurrence (Sharan et al, 2017; Cohen et al, 2009).3,4 Several biomarkers and therapeutic targets for OSCC have been demonstrated previously and may be useful for the diagnosis and prognosis of oral cancer in the future.5–9 Therefore, efforts are still needed to develop effective targeted therapies for OSCC.
Integrins are a family of transmembrane receptors; they form heterodimeric complexes composed of α and β subunits.10–13 The 18α and 8β subunits are members of approximately 24 different Integrin receptors, each of which is capable of binding to specific ligands. They function in specific signal transduction pathways and in cell–cell adhesion and the adhesion between cells and the ECM.14 Integrin signalling may be involved in proliferation, invasion and migration.15
Integrin subunit α5 (ITGA5) often combines with ITGB1 to form Integrin α5β1, which serves as a receptor for cell differentiation, cell development and migration.16,17 The emergence of Integrin α5β1 expression was found to be associated with tumour progression in lung cancer.18 In this study, we detected the expression levels of Integrin α5/ITGA5 in tissue and cell lines by qRT-PCR, immunohistochemistry and Western blotting and uncovered its biological function through a series of experiments.
Materials And Methods
Tissue Samples
All human tissues were obtained from the surgical suite in the Department of Stomatology at the First Affiliated Hospital of Sun Yat-sen University after identification by a pathologist. Tissues were obtained with the patients’ written and informed consent under a protocol approved by the institution’s Institutional Review Board. A total of 105 samples were taken. All the samples were taken from tongue.
Immunohistochemistry
Immunohistochemical analysis was performed to measure the level of Integrin α5/ITGA5. In brief, slides were rehydrated and blocked, and 7 slides were incubated with primary anti-Integrin α5/ITGA5 (1:100 dilution, Abcam) overnight at 4°C. After washing, slides were incubated with an anti-rabbit antibody conjugated to peroxidase streptavidin. A positive signal was detected with DAB (3,3ʹ-diaminobenzidine).
Cell Culture
The normal epithelial HaCaT cell line and the human OSCC cell lines CAL27, SCC-9 and SCC-25 were obtained from American Type Culture Collection (Manassas, VA, USA). Other cell lines were gifts from Professor Anxun Wang of the Department of Stomatology at the First Affiliated Hospital of Sun Yat-sen University approved by the First Affiliated Hospital of Sun Yat-sen University Research Committee. HaCaT and CAL27 cells were cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) with 10% foetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and penicillin–streptomycin, while other cell lines were cultured in DMEM F-12 medium containing 10% FBS at 37°C in a humidified tissue culture incubator with 5% CO2. All the cells were HPV free.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA from cells or tissues was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the protocol, and 2000 ng of RNA was used to obtain cDNA through reverse transcription by using PrimeScript RT Master Mix (Takara Bio, Kusatsu, Japan). Quantitative RT PCR (qRT-PCR) was conducted using SYBR Green PCR Master Mix (Takara Bio). Relative expression was determined by normalizing to the expression of GAPDH. The key primers are listed as in the following:
 |
|
Western Blot Assay
Equal amounts of protein extracts were separated via 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). After the membrane was blocked, a primary antibody (ITGA5 CST98204 PCNA CST13110 N-cad CST13116 E-cad CST14472 Vimentin CST5741 Snail CST3879 β-actin CST3700) was added for an overnight incubation at 4°C. Finally, the membrane was incubated with its respective horseradish peroxidase-conjugated secondary antibody (KPL 27856 27857) for 1 hr. The protein bands of interest were visualized using enhanced chemiluminescence reagents (Millipore, Burlington, MA, USA).
Colony Formation Assay
To measure colony formation, 2000 cells/well were seeded and incubated for 14 days to allow the formation of colonies. The colonies were fixed using methanol and stained with 0.1% crystal violet for 10 mins. The number of colony was counted through ImageJ.
Cell Counting Kit-8 (CCK-8) Assay
The indicated cells were seeded into 96-well plates at a density of 2×103 cells per well. Cell viability was determined using a CCK-8 assay (Dojindo, Kumamoto, Japan) every 24 hrs and by measuring absorbance at 450 nm following the manufacturer’s instructions.
Lentiviral Production And Stable Cell Line Construction
Lentiviral vectors expressing an shRNA or Integrin α5/ITGA5 (biochemistry Shanghai) were co-transfected with packaging vectors psPAX2 and pMD2G (Addgene) into HEK293FT cells for lentivirus production; the transfection was performed using Lipofectamine 3000 in accordance with the manufacturer’s instructions. To establish stable cell lines, cells were infected with lentiviruses as described above. After incubating for 72 hrs, cells were selected with 2 μg/mL puromycin treatment for 3 days. shRNA-1 target sequence AACAGAAAATAAAACAGA, shRNA-2 target sequence ACGAACCTCTTCTGTGATGGA.
Trans-Well Assay
Migration and invasion were assessed using trans-well plates. For the invasion assay, 5×104 cells were resuspended in 250 μL of plain medium without any FBS in the upper chamber (8-μm pore size, Costar, Corning, NY, USA) while the lower chamber was filled with 0.75 mL of complete medium with 10% FBS. After incubating for 2 days at the incubator, the upper chamber was fixed with 100% methanol and stained with 0.1% crystal violet. For the migration assay, 5×104 cells were plated on chambers. The transmembrane cells were estimated under a microscope (Nikon, Tokyo, Japan) at 200× magnification.
Wound Healing Assay
We applied wound healing assay to detect the migration ability of cells. We seeded 1×105 cells per well into the 6-well plate. After, the cells meet the 100% density. Equal wound was made, after incubating for certain time. The relative width was measured through ImageJ.
Edu Assay
Transfected cells were seeded into 96-well plates at a density of 500 cells per well, After incubating for 24 hrs. Edu was applied according to the manufactory’s protocol (Raybio 251453). Images were taken from Olympus FV100 microscopy with red light of fluorescence. All images were taken in 200× magnification, the percentage of Edu+ cells were calculated by the confocal automatically.
Statistical Analysis
Statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Data are presented as the mean±SD. The chi-squared test was used to analyse clinicopathological characteristics. Comparisons between two groups were evaluated using a Student’s t-test. Differences among the groups were tested using a one-way ANOVA. All experiments were repeated at least three times. A P-value <0.05 was considered statistically significant.
Results
ITGA5 Is Upregulated In OSCC
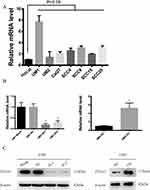
We scanned the TCGA database (519 OSCC tumours and 44 normal tissues) and identified ITGA5 as one of the most dysregulated genes between the two tissue types. To verify the results, we detected the mRNA level of ITGA5 in 105 OSCC tumours and paired normal tissues from the Department of Stomatology of the First Affiliated Hospital of Sun Yat-sen University by qRT-PCR. ITGA5 was upregulated in OSCC tumours from both the TCGA database and our own database (p<0.001) (Figure 1A and B). We further analysed the relationship between ITGA5 and clinical stages. Patients with an advanced stage harbour higher levels of ITGA5 than those patients at earlier stages (Figure 1C). To further confirm the results, we collected 10 paired tumour and normal tissues and used Western blotting to detect the protein levels of ITGA5. Corresponding to the mRNA level, the protein level of ITGA5 in tumours was higher than it was in normal tissues in 8 of 10 patients, as shown in Figure 1D.
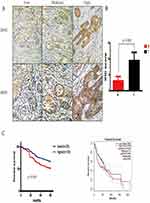
ITGA5 Is Correlated With OSCC Prognosis
We have proven that ITGA5 is upregulated in OSCC tissue. To uncover the relationship between ITGA5 and prognosis, we applied an IHC assay to detect the protein level of ITGA5 and analysed the IHC score. A representative image is shown in Figure 2A. The IHC score was higher in cancer tissue than it was in normal tissue (Figure 2B). We divided the patients into two groups. Both the TCGA database and our own database showed that patients with higher levels of ITGA5 had shorter overall survival times. Taken together, these data show that ITGA5 was negatively correlated with prognosis (Figure 2C), the characteristics of patients are shown in Table 1.
 |
Table 1 The Clinical Characteristic Of Patients |
Stable Cell Lines Were Successfully Constructed
To explore the biological function of ITGA5, we detected its mRNA levels in OSCC cell lines and found that OSCC cell lines harbour higher levels of ITGA5 than normal epithelium cells. Among the OSCC cell lines, UM1 had the highest level of ITGA5, while UM2 had the lowest level of ITGA5 (Figure 3A). Using UM1 cells, we established a stable knockdown cell line, and using UM2 cells we generated stable ITGA5-overexpressing cells. The mRNA and protein levels of ITGA5 in different cell lines were detected through qRT-PCR and Western blot experiments (Figure 3B and C).
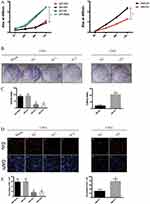
ITGA5 Promotes The Proliferation Of OSCC
To uncover the biological function of ITGA5 in OSCC, we used the CCK-8 assay, the colony formation assay and the EdU assay. The results showed that all the cells with higher levels of ITGA5 showed higher proliferation rates in both the UM1 and UM2 cell lines in the CCK-8 assay and the colony formation assay (Figure 4A–C). DZIP1-overexpressing cells harbour a high percentage of EdU-positive cells, while DZIP1-knockdown cells harbour a low percentage of Edu-positive cells (Figure 4D and E). Taken together, these results show that DZIP1 promotes the proliferation of OSCC in both short and long time periods.
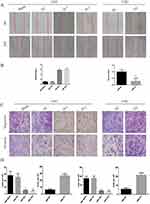
ITGA5 Promotes The Migration And Invasion Of OSCC
Recurrence and metastasis are two major factors responsible for OSCC-related death. We applied the wound healing assay, trans-well assay and invasion chamber assay to analyse the migration and invasion abilities of the cells described above. The cells with higher levels of ITGA5 migrated and invaded more easily than those with low ITGA5 levels. The wound healing assay showed greater motility of the UM1 control cells and UM2-OV cells than the other cells tested (Figure 5A and B). Cells with higher levels of ITGA5 migrate and invade more easily than other cells (Figure 5C and D).
ITGA5 Promotes The EMT Of OSCC
We have proven that ITGA5 serves as an oncogene and promotes the progression of OSCC by promoting proliferation, migration and invasion. EMT is an important process reflecting the invasive status of cancer cells. We detected EMT markers in the cell line described above. Mesenchymal markers such as N-cadherin, Vimentin and Snail were increased in cells with higher levels of ITGA5, but epithelial markers such as E-cadherin decreased at both the protein and mRNA levels, which corresponded to the former results (Figure 6A and B).
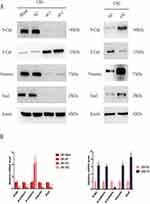 |
Figure 6 ITGA5 promotes the EMT of OSCC. (A) Western blot of EMT markers of OSCC cell line. (B) The relative mRNA level of EMT markers of OSCC cell line. *p<0.05. |
Discussion
OSCC is the most common malignancy in the Department of Stomatology, and current therapy is still surgical operation with adjuvant chemotherapy and radiotherapy. However, the overall survival time is unsatisfactory, and more biomarkers and therapeutic targets are needed.
We scanned the TCGA database and identified ITGA5 as one of the most dysregulated genes in OSCC. To verify that observation, we examined ITGA5 levels in 105 cancer tissues and paired normal tissues and assessed its correlation with prognosis. The results showed that ITGA5 was upregulated in cancer tissue and was negatively correlated with prognosis. To uncover the biological function of ITGA5 (specifically in proliferation and migration), we next measured the levels of ITGA5 and established stable knockdown and overexpression cell lines. The results showed that cells with higher levels of ITGA5 had higher proliferation, migration and invasion abilities compared to cells with lower ITGA5 levels. We next detected EMT markers and found that mesenchymal markers increased, while epithelial markers decreased with ITGA5 overexpression.
ITGA5 is a member of the Integrin family. Other family members, such as ITGA1/3, were reported to be upregulated in cancers and were correlated with cancer prognosis in cancers such as prostate cancer and breast cancer.19,20 Some research has suggested that ITGA5 is a potential biomarker in lung cancer. However, the biological function and underlying mechanisms of ITGA5 remain unclear. Integrin was reported to exert its function through various mechanisms. Wu showed that Integrin promotes invasion and metastasis via the PI3K and MAPK pathways.21 Integrin was able to promote the activation of the IL-1α pathway in both autocrine and paracrine manners.22 Integrin was reported to engage in the remodelling of the extracellular matrix and cell–cell adhesion in leukaemia.23 Studies have shown that Integrin promotes the production of reactive oxygen species (ROS), which protect cancer cells from apoptosis.24 Drugs or targeting chemicals were applied for high ROS levels and were recently reported as potential therapeutic targets.25 Taken together, these findings may provide insights into a potential therapeutic target in the future.
Conclusion
Integrin α5/ITGA5 promotes the proliferation, migration and invasion of oral squamous carcinoma by promoting EMT and may be a potential biomarker in the future.
Ethical Approval
All procedures performed that involve human participants were in accordance with the ethical standards of the First Affiliated Hospital of Sun Yat-sen University Research Committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards; studies were performed with the agreement of all patients.
Availability Of Data And Material
All data and material in this article are available, and the original version will be available upon demand.
Acknowledgment
We thank Professor Anxun Wang for excellent technical support.
Author Contributions
Yan designed the research. All authors contributed to data analysis, drafting or revising the article, gave approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declare no conflicts of interest.
References
1. Adachi M, Taki T, Higashiyama M, et al. Significance of integrin alpha5 gene expression as a prognostic factor in node-negative non-small cell lung cancer. Clin Cancer Res. 2000;6(1):96–101.
2. Adams C, Johnson S. Global health and cancer. Lancet. 2019;393(10175):984. doi:10.1016/S0140-6736(19)30010-8
3. Bromberger T, Klapproth S, Rohwedder I, et al. Direct Rap1/Talin1 interaction regulates platelet and neutrophil integrin activity in mice. Blood. 2018;132(26):2754–2762. (c) 2018 by The American Society of Hematology. doi:10.1182/blood-2018-04-846766.
4. Chen TW, Lee CC, Liu H, et al. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat Commun. 2017;8(1):465. doi:10.1038/s41467-017-00493-9
5. Cohen EE, Davis DW, Karrison TG, et al. Erlotinib and bevacizumab in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck: a phase I/II study. Lancet Oncol. 2009;10(3):247–257. doi:10.1016/S1470-2045(09)70002-6
6. Cooper J, Giancotti FG. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell.2019;35(3):347–367. doi:10.1016/j.ccell.2019.01.007
7. Das L, Anderson, TA, Gard JM, et al. Characterization of laminin binding integrin internalization in prostate cancer cells. J Cell Biochem. 2017;118(5):1038–1049. doi:10.1002/jcb.25673.
8. Dayan A, Fleminger G, Ashur-Fabian O. Targeting the Achilles’ heel of cancer cells via integrin-mediated delivery of ROS-generating dihydrolipoamide dehydrogenase. Oncogene. 2019;38:5050–5061. doi:10.1038/s41388-019-0775-9
9. Ginsberg MH. Integrin activation. BMB Rep. 2014;47(12):655–659. doi:10.5483/BMBRep.2014.47.12.241
10. Horvath J, Szabo A, Tar I, et al. Oral health may affect the performance of mRNA-based saliva biomarkers for oral squamous cell cancer. Pathol Oncol Res. 2018;24(4):833–842. doi:10.1007/s12253-017-0296-1
11. Hsu E-C, Kulp SK, Huang H-L, et al. Integrin-linked kinase as a novel molecular switch of the IL-6-NF-κB signaling loop in breast cancer. Carcinogenesis. 2016;37(4):430–442. doi:10.1093/carcin/bgw020.
12. Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69(1):11–25. doi:10.1016/0092-8674(92)90115-S
13. Li B-P, Mao Y-T, Wang Z, et al. CLIC1 promotes the progression of gastric cancer by regulating the MAPK/AKT pathways. Cell Physiol Biochem. 2018;46(3):907–924. doi:10.1159/000488822.
14. Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi:10.1136/gutjnl-2013-305806.
15. Moreno-Layseca P, Icha J, Hamidi H, Ivaska J. Integrin trafficking in cells and tissues. Nat Cell Biol. 2019;21(2):122–132. doi:10.1038/s41556-018-0223-z
16. Mostafavi-Pour Z, Ashrafi MR, Talaei-Khozani T. Down regulation of ITGA4 and ITGA5 genes after formation of 3D spherules by human Wharton’s jelly stem cells (hWJSCs). Mol Biol Rep. 2018;45(3):245–252. doi:10.1007/s11033-018-4157-0
17. Ren JS, Kamangar F, Qiao YL, et al. Serum pepsinogens and risk of gastric and oesophageal cancers in the general population nutrition intervention trial cohort. Gut. 2009;58(5):636–642. doi:10.1136/gut.2008.168641
18. Rivera C, Oliveira AK, Costa RAP, De Rossi T, Paes Leme AF. Prognostic biomarkers in oral squamous cell carcinoma: a systematic review. 2017. Oral Oncol. 72:38–47.
19. Ross MH, Esser AK, Fox GC, et al. Bone-induced expression of integrin β3 enables targeted nanotherapy of breast cancer metastases. Cancer Res. 2017;77(22):6299–6312. doi:10.1158/0008-5472.CAN-17-1225.
20. Schnittert J, Bansal R, Storm G, Prakash J. Integrins in wound healing, fibrosis and tumor stroma: high potential targets for therapeutics and drug delivery. Adv Drug Deliv Rev.2018;129:37–53. doi:10.1016/j.addr.2018.01.020
21. Sharan SS, Kumar R, Singh KV, et al. Expression of radioresistant gene PEG10 in OSCC patients and its prognostic significance. Asian Pac J Cancer Prev. 2017;6:1513–1518. Creative Commons Attribution License.
22. Sun Z, Costell M, Fassler R. Integrin activation by talin, kindlin and mechanical forces. Nat Cell Biol. 2019;21(1):25–31. doi:10.1038/s41556-018-0234-9
23. Varkas G, Elewaut D. Response to: ‘Simultaneous inhibition of α4/β7 integrin and tumor necrosis factor-α in concomitant spondyloarthritis and inflammatory bowel disease’ by Richard et al. Ann Rheum Dis. 2018;77(12):e87. doi:10.1136/annrheumdis-2017-212858
24. Yazlovitskaya EM, Viquez OM, Tu T, et al. The laminin binding alpha3 and alpha6 integrins cooperate to promote epithelial cell adhesion and growth. Matrix Biol. 2018;77:101–116.
25. Zhao F, Qiao Y. Cervical cancer prevention in China: a key to cancer control. Lancet. 2019;393(10175):969–970. doi:10.1016/S0140-6736(18)32849-6
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.